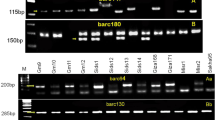
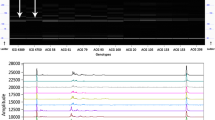
Abstract
Cultivated peanut, the second most economically important legume crop throughout the United States and the third most important oilseed in the world, is consistently threatened by various diseases and pests. Sclerotinia minor Jagger (S. minor), the causal agent of Sclerotinia blight, is a major threat to peanut production in the Southwestern U.S., Virginia, and North Carolina and can reduce yield by up to 50% in severely infested fields. Although host plant resistance would provide the most effective solution to managing Sclerotinia blight, limited sources of resistance to the disease are available for use in breeding programs. Peanut germplasm collections are available for exploration and identification of new sources of resistance, but traditionally the process is lengthy, requiring years of field testing before those potential sources can be identified. Molecular markers associated with phenotypic traits can speed up the screening of germplasm accessions, but until recently none were available for Sclerotinia blight resistance in peanut. This study objective of this study was to characterize the US peanut mini-core collection with regards to a recently discovered molecular marker associated with Sclerotinia blight resistance. Ninety-six accessions from the collection were available and genotyped using the SSR marker and 39 total accessions from spanish, valencia, runner market types were identified as new potential sources of resistance and targeted for further evaluation in field tests for Sclerotinia blight resistance.

Similar content being viewed by others
References
Baring MR, Simpson CE, Burow MD, Black MC, Cason JM, Ayers J, Lopez Y, Melouk HA (2006) Registration of ‘Tamnut OL07’ peanut. Crop Sci 46:2721–2722
Barkley NA, Dean R, Pittman RN, Wang ML, Holbrook CC, Pederson GA (2007) Genetic diversity of cultivated and wild-type peanuts evaluated with M13-tailed SSR markers and sequencing. Genet Res Camb 89:93–106
Chappell GF, Shew BB, Ferguson JM, Beute MK (1995) Mechanisms of resistance to Sclerotinia minor in selected peanut genotypes. Crop Sci 35:692–696
Chenault KD, Gallo M, Seib JC, James VA (2007) A non-destructive seed sampling method for PCR-based analyses in marker-assisted selection and transgene screening. Peanut Sci 34:38–43
Chenault KD, Maas A, Melouk HA, Payton ME (2008) Discovery and characterization of a molecular marker for Sclerotinia minor (Jagger) resistance in peanut. Euphytica. doi:10.1007/s10681-008-9816-0
Chu Y, Holbrook CC, Timper P, Ozais-Akins P (2007a) Development of a PCR-based molecular marker to select for nematode resistance in peanut. Crop Sci 47:841–847
Chu Y, Ramos L, Holbrook CC, Ozais-Akins P (2007b) Frequency of a loss-of-function mutation in oleoyl-PC desaturase (ahFAD2A) in the mini core of the U.S. peanut germplasm collection. Crop Sci 47:2372–2378
Coffelt TA, Porter DM (1982) Screening peanuts for resistance to Sclerotinia blight. Plant Dis 71:811–815
Coyne DP, Steadman JR, Anderson FN (1974) Effect of modified plant architecture of great northern dry bean varieties (Phaseolus vulgaris) on white old severity, and components of yield. Plant Dis Rep 58:379–382
He G, Prakash CS (1997) Identification of polymorphic DNA markers in cultivated peanut (Arachis hypogaea L.). Euphytica 97:143–149
He G, Ronghua M, Gao H, Guo B, Gao G, Newman M, Pittman RN, Prakash CS (2005) Simple sequence repeat markers for botanical varieties of cultivated peanut (Arachis hypogaea L.). Euphytica 142:131–136
Holbrook CC, Anderson WF (1995) Evaluation of a core collection to identify resistance to late leafspot in peanut. Crop Sci 35:1700–1702
Holbrook CC, Dong W (2005) Development and evaluation of a mini core collection for the U.S. peanut germplasm collection. Crop Sci 45:1540–1544
Holbrook CC, Anderson WF, Pittman RN (1993) Selection of a core collection from the U.S. germplasm collection of peanut. Crop Sci 33:859–861
Hopkins MS, Casa AM, Wang T, Mitchell SE, Dean RE, Kochert GD, Kresovich S (1999) Discovery and characterization of polymorphic simple sequence repeats (SSRs) in cultivated peanut (Arachis hypogaea L.). Crop Sci 39:1243–1247
Kang I, Gallo M, Tillman BL (2007) Distribution of allergen composition in peanut (Arachis hypogaea L) and wild progenitor (Arachis) species. Crop Sci 47:997–1003
Kochert G, Halward TM, Branch WD, Simpson CE (1991) RFLP variability in peanut cultivars and wild species. Theor Appl Genet 81:565–570
Kottapalli KR, Burow MD, Burow G, Burke J, Puppala N (2007) Molecular characterization of the U.S. peanut mini core collection using microsatellite markers. Crop Sci 47:1718–1727
Melouk HA, Backman PA (1995) In Melouk HA, Shokes FM (eds) Peanut health management. APS Press, St. Paul, MN, pp 75–85
Schwartz HF, Steadman JR, Coyne DP (1978) Influence of Phaseolus vulgaris blossoming characteristics and canopy structure upon reaction to Sclerotinia sclerotiorum. Phytopathology 68:465–470
Simpson CE, Krapovickas A, Valls JM (2001) History of Arachis including evidence of A. hypogaea progenitors. Peanut Sci 28:79–81
Smith OD, Simpson CE, Grichar WJ, Melouk HA (1991) Registration of ‘Tamspan 90’ peanut. Crop Sci 31:1711
Smith OD, Simpson CE, Black MC, Besler BA (1998) Registration of ‘Tamrun 96’ peanut. Crop Sci 38:1403
Stalker HT, Mozingo LG (2001) Molecular markers of Arachis and marker-assisted selection. Peanut Sci 28:117–123
Upadhyaya HD, Bramel PJ, Ortiz R, Singh S (2002) Developing a mini core of peanut for utilization of genetic resources. Crop Sci 42:2150–2156
Wildman LG, Smith OD, Simpson CE, Taber RA (1992) Inheritance of resistance to Sclerotinia minor in selected Spanish peanut crosses. Peanut Sci 19:31–34
Acknowledgments
The authors would like to thank Shannon Stoup, Lisa Myers and Doug Glasgow for their technical assistance involved in this research. Appreciation should also be given to Dr. Corley Holbrook of USDA-ARS in Tifton, GA, and to Dr. Roy Pittman, curator of the USDA-ARS peanut germplasm collection located in Griffin, GA, for supplying germplasm accessions for analysis. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chenault Chamberlin, K.D., Melouk, H.A. & Payton, M.E. Evaluation of the U.S. peanut mini core collection using a molecular marker for resistance to Sclerotinia minor Jagger. Euphytica 172, 109–115 (2010). https://doi.org/10.1007/s10681-009-0065-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-009-0065-7