Abstract
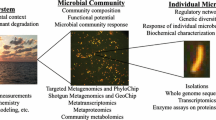
This article brings forth recommendations from a workshop sponsored by the U.S. Environmental Protection Agency's Science to Achieve Results (STAR) and Environmental Monitoring and Assessment (EMAP) Programs and by the Council of State Governments, held during May 2002 in Kansas City, Kansas. The workshop assembled microbial ecologists and environmental scientists to determine what research and science is needed to bring existing molecular biological approaches and newer technologies arising from microbial genomic research into environmental monitoring and water quality assessments. Development of genomics and proteomics technologies for environmental science is a very new area having potential to improve environmental water quality assessments. The workshop participants noted that microbial ecologists are already using molecular biological methods well suited for monitoring and water quality assessments and anticipate that genomics-enabled technologies could be made available for monitoring within a decade. Recommendations arising from the workshop include needs for (i) identification of informative microbial gene sequences, (ii) improved understandings of linkages between indicator taxa, gene expression and environmental condition, (iii) technological advancements towards field application, and (iv) development of the appropriate databases.
Similar content being viewed by others
References
Adamczyk, J., Hesselsoe, M., Iversen, N., Horn, M., Lehner, A., Nielsen, P. H., Schloter, M., Roslev, P. and Wagner, M.: 2003, ‘The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function’, Appl. Environ. Microbiol. 69, 6875–6887.
Amann, R. I., Ludwig, W. and Schleifer, K. H.: 1995, ‘Phylogenetic identification and in situ detection of individual microbial cells without cultivation’, Microbiol. Rev. 59, 143–169.
Barkay, T., Fouts, D. L. and Olson, B. H.: 1985, ‘Preparation of a DNA gene probe for detection of mercury resistance in Gram-negative bacteria’, Appl. Environ. Microbiol. 49, 1196–1202.
Bavykin, S. G., Akowski, J. P., Zakhariev, V. M., Barsky, V. E., Perov, A. N. and Mirzabekov, A. D.: 2001, ‘Portable system for microbial sample preparation and oligonucleotide microarray analysis’, Appl. Environ. Microbiol. 67, 922–928.
Beja, O., Suzuki, M. T., Koonin, E. V., Aravind, L., Hadd, A., Nguyen, L. P., Vilacorta, R., Amjadi, M., Garrigues, C., Jovanovich, S. B., Feldman, R. A. and DeLong, E. F.: 2000, ‘Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage’, Environ. Microbiol. 2, 516–529.
Beja, O., Spudich, E. N., Spudich, J. L., LeClerc, M. and DeLong, E. F.: 2001, ‘Proteorhodopsin phototrophy in the ocean’, Nature 411, 786–789.
Berggren, C., Stålhandske, P., Brundell, J. and Johansson, G: 1999, ‘A feasibility study of a capacitive biosensor for direct detection of DNA hybridization’, Electroanalysis 11, 156–160.
Bernhard, A. E. and Field, K. G.: 2000a, ‘Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes’, Appl. Environ. Microbiol. 66, 1587–1594.
Bernhard, A. E. and Field, K. G.: 2000b, ‘A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA’, Appl. Environ. Microbiol. 66, 4571–4574.
Chen, F., Gonzalez, J. M., Dustman, W. A., Moran, M. A. and Hodson, R. E.: 1997, ‘In situ reverse transcription, an approach to characterize genetic diversity and activities of prokaryotes’, Appl. Environ. Microbiol. 63, 4907–4913.
Cho, J.-C. and Tiedje, J. M.: 2002, ‘Quantitative detection of microbial genes by using DNA microarrays’, Appl. Environ. Microbiol. 68, 1425–1430.
Connon, S. A. and Giovannoni, S. J.: 2002, ‘High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates’, Appl. Environ. Microbiol. 68, 3878–3885.
DeLeon, R., Shieh, Y. S. C., Baric, R. S. and Sobsey, M. D.: 1990, ‘Detection of enteroviruses and hepatitis A virus in environmental samples by gene probes and polymerase chain reaction’, in: Proceeding of the Water Quality Conference, San Diego, CA, American Water Works Association, Vol. 18, Denver, CO., pp. 833–853.
Derisi, J. L., Iyer, V. R. and Brown, P. O.: 1997, ‘Exploring the metabolic and genetic control of gene expression on a genomic scale’, Science 278, 680–686.
Dick, L. K. and Field, K. G.: 2004, ‘Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes’, Appl. Environ. Microbiol. 70, 5696–5697.
Fan, C., Li, G., Gu, Q., Zhu, J. and Zhu, D.: 2000, Electrochemical detection of Cecropin CM4 gene by single stranded probe and cysteine modified gold electrode, Anal. Lett. 33, 1479–1490.
Ferris, M. J. and Ward, D. M.: 1997, ‘Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis’, Appl. Environ. Microbiol. 63, 1375–1381.
Fisher, W. S., Jackson, L. E. and Kurtz, J. C.: 2005, ‘U.S. EPA Office of Research and Development guidelines for technical evaluation of ecological indicators’, in: D. J. Rapport, W.L, Lasley, D. E. Rolston, N. O. Nielsen, C. O. Qualset, A. B. Damania (eds), Managing for Healthy Ecosystems, Lewis CRC Press, Boca Raton, FL, pp. 277–284.
Fox, J. D., Han, S., Samuelson, A., Zhang, Y., Neale, M. L. and Westmoreland, D.: 2002, ‘Development and evaluation of nucleic acid sequence based amplification (NASBA) for diagnosis of enterovirus infections using the NucliSens®, Basic Kit’, J. Clin. Virol. 24, 117–130.
Fuhrman, J. A., Griffith, J. F. and Schwalbach, M. S.: 2002, ‘Prokaryotic and viral diversity patterns in marine plankton’, Ecol. Res. 17, 183–194.
Girones, R., Puig, M., Allard, A., Lucena, F., Wadell, G. and Jofre, J.: 1995, ‘Detection of adenovirus and enterovirus by PCR amplification in polluted waters’, Water Sci. Technol. 31, 351–357.
Griffin, T. J., Goodlett, D. R. and Aebersold, R.: 2001a, ‘Advances in proteome analysis by mass spectrometry’, Curr. Opin. Biotechnol. 12, 607–612.
Griffin, T. J., Gygi, S. P., Rist, B., Aebersold, R., Loboda, A., Jilkine, A., Ens, W. and Standing, K. G.: 2001b, ‘Quantitative proteomic analysis using a MALDI quadrupole time-of-flight mass spectrometer’, Anal. Chem. 73, 978–986.
Gruntzig, V., Nold, S. C., Zhou, J. and Tiedje, J. M.: 2001, ‘Pseudomonas stutzeri nitrite reductase gene abundance in environmental samples measured by real-time PCR’, Appl. Environ. Microbiol. 67, 760–768.
Haugland, R. A., Siefring, S. C., Wymer, L. J., Brenner, K. P. and Dufour, A. P.: 2005, ‘Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis’, Water Res. 39, 559–568.
Huber, M., Losert, D., Hiller, R., Harwanegg, C., Mueller, M. W. and Schmidt, W. M.: 2001, ‘Detection of single base alterations in genomic DNA by solid phase polymerase chain reaction on oligonucelide microarrays’, Anal. Biochem. 299, 24–30.
Jiang, M. and Wang, J.: 2001, ‘Recognition and detection of oligonucleotides in the presence of chromosomal DNA based on entrapment within conducting-polymer networks’, J. Electroanal. Chem. 500, 584–589.
Kerkhof, L., Santoro, M. and Garland, J.: 2000, ‘Response of soybean rhizosphere communities to human hygiene water addition as determined by community level physiological profiling (CLPP) and terminal restriction fragment length polymorphism (TRFLP) analysis’, FEMS Microbiol. Lett. 18, 95–101.
Koizumi, Y., Kelly, J. J., Nakagawa, T., Urakawa, H., El-Fantroussi, S., Al-Muzaini, S., Fukui, M., Urushigawa, Y. and Stahl, D. A.: 2002, ‘Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology’, Appl. Environ. Microbiol. 68, 3215–3225.
Kozwich, D., Johansen, K. A., Landau, K., Roehl, C. A., Woronoff, S. and Roehl, P. A.: 2000, ‘Development of a novel, rapid integrated Cryptosporidium parvum detection assay’, Appl. Environ. Microbiol. 66, 2711–2717.
Liu, W.-T., Marsh, T. L., Cheng, H. and Forney, L. J.: 1997, ‘Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA’, Appl. Environ. Microbiol. 63, 4516–4522.
Liu, W. T., Mirzabekov, A. D. and Stahl, D. A.: 2001, ‘Optimization of an oligonucleotide microchip for microbial identification studies: A non-equilibrium dissociation approach’, Environ Microbiol. 3, 619–629.
Lucchini, S., Thompson, A. and Hinton, J. C.D.: 2001, ‘Microarrays for microbiologists’, Microbiology 147, 1403–1414.
Marrazza, G., Chianell, I. and Mascin, M.: 1999, ‘Disposable DNA electrochemical biosensors for environmental monitoring’, Anal. Chim. Acata. 387, 297–307.
Marsh, T. L., Saxman, P., Cole, J. and Tiedje, J. M.: 2000, ‘Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis’, Appl. Environ. Microbiol. 66, 3616–3620.
Morris, R. M., Rappé, M. S., Connon, S. A., Vergin, K. L., Siebold, W. A., Carlson, C. A. and Giovannoni, S. J.: 2002, ‘SAR11 clade dominates ocean surface bacterioplankton communities’, Nature 420, 806–810.
Moyer, C. L.: 2001, ‘Molecular phylogeny’, in: J. H. Paul (ed.), Marine Microbiology, Meth. Microbiol. 30, 375–394.
Muyzer, G., de Waal, E. C. and Uitterlinden, A. G.: 1993, ‘Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA’, Appl. Environ. Microbiol. 59, 695–700.
National Science and Technology Council: 2001, ‘The Microbe Project: A report from the Interagency Working Group on Microbial Genomics’ (http://www.ostp.gov/html/microbial/ pdf_files/microbe.pdf).
Ogunseitan, O. A.: 1997, ‘Direct extraction of catalytic proteins from natural microbial communities’, J. Microbiol. Meth. 28, 55–63.
Ogunseitan, O. A.: 2000, ‘Microbial proteins as biomarker of ecosystem health’, in: K. M. Scow, G.E Fogg, D. E. Hinton and M. L. Johnson (eds), Integrated Assessment of Ecosystem Health, CRC Press, Boca Raton, FL, pp. 207–222.
Ogunseitan, O. A., Yang, S. and Ericson, J.: 2000, ‘Microbial δ-aminolevulinate dehydratase as a biosensor of lead bioavailability in contaminated environments’, Soil Biol. Biochem. 32, 1899–1906.
Olsen, G. J., Lane, D. J., Giovannoni, S. J. and Pace, N. R.: 1986, ‘Microbial ecology and evolution: A ribosomal RNA approach’, Annu. Rev. Microbiol. 40, 337–365.
Ouverney, C. C. and Fuhrman, J. A.: 2000, ‘Marine planktonic Archaea take up amino acids’, Appl. Environ. Microbiol. 66, 4829–4833.
Overbeck, J. and Chrost, R.J (eds): 1990. Aquatic Microbial Ecology: Biochemical and Molecular Approaches, Springer-Verlag, New York.
Pace, N. R., Stahl, D. A., Lane, D. J. and Olsen, G. J.: 1986, ‘The analysis of natural microbial populations by ribosomal RNA sequences’, Adv. Microbiol. Ecol. 9, 1–55.
Paul, J. H., Pichard, S. L., Kang, J. B., Watson, G. M. F. and Tabita, F. R.: 1999, ‘Evidence for a clade-specific temporal and spatial separation in ribulose bisphosphate carboxylase gene expression in phytoplankton populations off Cape Hatteras and Bermuda’, Limnol. Oceanogr. 44, 12–23.
Peplies, J., Lau, S. C. K., Pernthanler, J., Amann, R. I. and Glöckner, F. O.: 2004, ‘Application and validation of DNA microarrays for the 16S rRNA-based analysis of marine bacterioplankton’, Environ. Microbiol. 6, 638–645.
Pernthaler, A., Pernthaler, J., Shattenhofer, M. and Amann, R. I.: 2002, ‘Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton’, Appl. Environ. Microbiol. 68, 5728–5736.
Rappé, M. S., Connon, S. A., Vergin, K. L. and Giovannoni, S. J.: 2002, ‘Cultivation of the ubiquitous SAR11 marine bacterioplankton clade’, Nature 418, 630–633.
Rose, J. B. and Grimes, J. D.: 2001, ‘Revaluation of microbial Water Quality: Powerful New Tools for Detection and Risk Assessment, A report from the American Academy of Microbiology’, American Society for Microbiology, Washington, DC.
Rose, J. B., Zhou, X., Griffin, D. W. and Paul, J. H.: 1997, ‘Comparison of PCR and plaque assay for detection and enumeration of coliphage in polluted marine waters’, Appl. Environ. Microbiol. 63, 4564–4566.
Rudi, K., Skulberg, O. M. and Jakobsen, K. S.: 2000, ‘Application of sequence-specific labeled 16S rRNA gene oligonucleotide probes for genetic profiling of cyanobacterial abundance and diversity by array hybridization’, Appl. Environ. Microbiol. 66, 4004–4011.
Santo Domingo, J. W., Siefring, S. C. and Haugland, R. A.: 2003, ‘Real-time PCR method to detect Enterococcus faecalis in water’, Biotechnol. Lett. 25, 261–265.
Schafer, H. and Muyzer, G.: 2001, ‘Denaturing gradient gel electrophoresis in marine microbial ecology’, in: J. H. Paul (ed.), Marine Microbiology, Meth. Microbiol. 30, 425–468.
Schena, M., Shalon, D., Davis, R. W. and Brown, P. O.: 1995, ‘Quantitative monitoring of gene expression patterns with a complementary DNA microarray’, Science 270, 467–470.
Short, R. T., Fries, D. P., Kerr, M. L., Lembke, C. E., Toler, S. K., Wenner, P. G. and Byrne, R. H.: 2001, ‘Underwater mass spectrometers for in situ chemical analysis of the hydrosphere’, J. Am. Soc. Mass Spectrom. 12, 676–682.
Small, J., Call, D. R., Brockman, F. J., Straub, T. M. and Chandler, D. P.: 2001, ‘Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays’, Appl. Environ. Microbiol. 67, 4708–4716.
Stahl, D. A. and Tiedje, J. M.: 2002, ‘Microbial Ecology and Genomics: A Crossroads of Opportunity’, A Report from the American Academy of Microbiology, ASM Press, Washington DC.
Staley, J. T., Castenholtz, R. W., Colwell, R. R., Holt, J. G., Kane, M. D., Pace, N. R., Salyers, A. A. and Tiedje, J. M.: 1997, ‘The Microbial World: Foundation of the Biosphere’, A Report from the American Academy of Microbiology, ASM Press, Washington, DC.
Straub, T. M., Daly, D. S., Wunshel, S., Rochelle, P. A., DeLeon, R. and Chandler, D. P.: 2002, ‘Genotypeing Cryptosporidium parvum with an hsp70 single-nucleotide polymorphism microarray’, Appl. Environ. Microbiol. 68, 1817–1826.
Suzuki, M. T., Taylor, L. T. and DeLong, E. F.: 2000, ‘Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays’, Appl. Environ. Microbiol. 66, 4605–4614.
Thompson, D. K., Beliaev, A. S., Giometti, C. S., Tollaksen, S. L., Khare, T., Lies, D. P., Nealson, K. H., Lim, H., Yates III, J., Brandt, C. C., Tiedje, J. M. and Zhou, J.: 2002, ‘Transcriptional and proteomic analysis of a ferric uptake regulator (Fur) mutant of Shewanella oneidensis: Possible involvement of Fur in energy metabolism, transcriptional regulation, and oxidative stress’, Appl. Environ. Microbiol. 68, 881–892.
United States Environmental Protection Agency: 1990, ‘Biological Criteria: National Program Guidance for Surface Waters’ (EPA-440/5-90-004).
Urakawa, H., Noble, P. A., El Fantroussi, S., Kelly, J. J. and Stahl, D. A.: 2002, ‘Single-base-pair discrimination of terminal mismatches by using oligonucleotide microarrays and neural network analyses’, Appl Environ Microbiol. 68, 235–244.
Wawrik, B., Paul, J. H. and Tabita, F. R.: 2002, ‘Real-Time PCR quantification of rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase) mRNA in diatoms and pelagophytes’, Appl. Environ. Microbiol. 68, 3771–3779.
Wu, L., Thompson, D. K., Li, G., Hurt, R. A., Tiedje, J. M. and Zhou, J.: 2001, ‘Development and evaluation of functional gene arrays for detection of selected genes in the environment’, Appl. Environ. Microbiol. 67, 5708–5790.
Ye, R. W., Wang, T., Bedzyk, L. and Coker, K. M.: 2001, ‘Applications of DNA microarrays in microbial systems’, J. Microbiol. Meth. 47, 257–272.
Zehr, J. P. and Turner, P. J.: 2001, ‘Nitrogen fixation: Nitrogenase genes and gene expression’, in: J. H. Paul (ed.), Marine Microbiology, Meth. Microbiol. 30, 271–289.
Author information
Authors and Affiliations
Corresponding author
Additional information
Contribution no. 1217 from the NHEERL Gulf Ecology Division.
All authors contributed equally to this publication.
Rights and permissions
About this article
Cite this article
Devereux, R., Rublee, P., Paul, J.H. et al. Development and Applications of Microbial Ecogenomic Indicators for Monitoring Water Quality: Report of a Workshop Assessing the State of the Science, Research Needs and Future Directions. Environ Monit Assess 116, 459–479 (2006). https://doi.org/10.1007/s10661-006-7665-7
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s10661-006-7665-7