Abstract
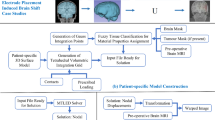
In this paper we evaluate the accuracy of warping of neuro-images using brain deformation predicted by means of a patient-specific biomechanical model against registration using a BSpline-based free form deformation algorithm. Unlike the BSpline algorithm, biomechanics-based registration does not require an intra-operative MR image which is very expensive and cumbersome to acquire. Only sparse intra-operative data on the brain surface is sufficient to compute deformation for the whole brain. In this contribution the deformation fields obtained from both methods are qualitatively compared and overlaps of Canny edges extracted from the images are examined. We define an edge based Hausdorff distance metric to quantitatively evaluate the accuracy of registration for these two algorithms. The qualitative and quantitative evaluations indicate that our biomechanics-based registration algorithm, despite using much less input data, has at least as high registration accuracy as that of the BSpline algorithm.










Similar content being viewed by others
Notes
IRB approval was acquired for the use of the anonymised retrospective image database for this study.
References
ABAQUS, ABAQUS Theory Manual Version 5.2. 1998, Hibbit, Karlsson & Sorensen, Inc.
Black, P. Management of malignant glioma: role of surgery in relation to multimodality therapy. J. Neurovirol. 4:227–236, 1998.
Box, G. E. P., W. G. Hunter, and J. S. Hunter. Statistics for Experimenters. An introduction to Design, Data Analysis, and Model Building. New York: John Wiley & Sons, 1978.
Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 8:679–698, 1986.
Fedorov, A., E. Billet, M. Prastawa, G. Gerig, A. Radmanesh, S. K. Warfield, R. Kikinis, and N. Chrisochoides. Evaluation of brain MRI alignment with the Robust Hausdorff distance measures. In: Advances in Visual Computing, Pt I, edited by G. Bebis. 2008, pp. 594–603.
Garlapati, R. R., G. Joldes, A. Wittek, J. Lam, N. Weisenfeld, A. Hans, S. K. Warfield, R. Kikinis, and K. Miller. Objective evaluation of accuracy of intra-operative neuroimage registration. In: Computational Biomechanics for Medicine VII: Models, Algorithms and Implementations, edited by A. Wittek, K. Miller, and P. M. F. Nielsen. New York: Springer, 2013, pp. 87–99.
Grosland, N. M., K. H. Shivanna, V. A. Magnotta, N. A. Kallemeyn, N. A. DeVries, S. C. Tadepalli, and C. Lislee. IA-FEMesh: an open-source, interactive, multiblock approach to anatomic finite element model development. Comput. Methods Programs Biomed. 94:96–107, 2009.
Hill, D. L. G. and P. Batchelor. Registration methodology: concepts and algorithms. In: Medical Image Registration, edited by J. V. Hanjal, D. L. G. Hill, and D. J. Hawkes. CRC Press, 2001, pp. 39–70.
Huttenlocher, D. P., G. A. Klanderman, and W. J. Rucklidge. Comparing images using the Hausdorff distance. IEEE Trans. Pattern Anal. Mach. Intell. 15:850–863, 1993.
Ji, S. B., X. Y. Fan, D. W. Roberts, and K. D. Paulsen. Cortical surface strain estimation using stereovision. In: Medical Image Computing and Computer-Assisted Intervention, MICCAI 2011, Pt I, edited by G. Fichtinger, A. Martel, and T. Peters. 2011, pp. 412–419.
Ji, S. B., D. W. Roberts, A. Hartov, and K. D. Paulsen. Brain-skull contact boundary conditions in an inverse computational deformation model. Med. Image Anal. 13:659–672, 2009.
Jin, X., G. R. Joldes, K. Miller, K. H. Yang, and A. Wittek. Meshless algorithm for soft tissue cutting in surgical simulation. Comput. Methods Biomech. Biomed. Eng. 2012. doi:10.1080/10255842.2012.716829
Jin, J. Y., F. F. Yin, S. E. Tenn, P. M. Medin, and T. D. Solberg. Use of the BrainLAB exactrac X-ray 6D system in image-guided radiotherapy. Med. Dosim. 33:124–134, 2008.
Joldes, G. R., A. Wittek, M. Couton, S. K. Warfield, and K. Miller. Real-time prediction of brain shift using nonlinear finite element algorithms. In: Medical Image Computing and Computer-Assisted Intervention—Miccai 2009, Pt II, edited by G. Z. Yang, D. Hawkes, D. Rueckert, A. Nobel, and C. Taylor. Berlin/Heidelberg: Springer, 2009, pp. 300–307.
Joldes, G. R., A. Wittek, and K. Miller. Computation of intra-operative brain shift using dynamic relaxation. Comput. Meth. Appl. Mech. Eng. 198:3313–3320, 2009.
Joldes, G. R., A. Wittek, and K. Miller. Non-locking tetrahedral finite element for surgical simulation. Commun. Numer. Methods Eng. 25:827–836, 2009.
Joldes, G. R., A. Wittek, and K. Miller. Suite of finite element algorithms for accurate computation of soft tissue deformation for surgical simulation. Med. Image Anal. 13:912–919, 2009.
Joldes, G. R., A. Wittek, and K. Miller. Cortical surface motion estimation for brain shift prediction. In: Computational Biomechanics for Medicine, edited by K. Miller, and P. M. F. Nielsen. New York: Springer, 2010, pp. 53–62.
Joldes, G. R., A. Wittek, and K. Miller. Real-time nonlinear finite element computations on GPU—application to neurosurgical simulation. Comput. Meth. Appl. Mech. Eng. 199:3305–3314, 2010.
Joldes, G. R., A. Wittek, and K. Miller. An adaptive dynamic relaxation method for solving nonlinear finite element problems. Application to brain shift estimation. Int. J. Numer. Methods Biomed. Eng. 27:173–185, 2011.
Joldes, G., A. Wittek, K. Miller, and L. Morriss. Realistic and efficient brain-skull interaction model for brain shift computation. In: Computational Biomechanics for Medicine III, edited by K. Miller, and P. M. F. Nielsen. New York: Springer, 2008.
Klein, A., J. Andersson, B. A. Ardekani, J. Ashburner, B. Avants, M.-C. Chiang, G. E. Christensen, D. L. Collins, J. Gee, P. Hellier, J. H. Song, M. Jenkinson, C. Lepage, D. Rueckert, P. Thompson, T. Vercauteren, R. P. Woods, J. J. Mann, and R. V. Parsey. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 46:786–802, 2009.
Kybic, J., P. Thevenaz, A. Nirkko, and M. Unser. Unwarping of unidirectionally distorted EPI images. IEEE Trans. Med. Imaging 19:80–93, 2000.
Kybic, J., and M. Unser. Fast parametric elastic image registration. IEEE Trans. Image Process. 12:1427–1442, 2003.
Lee, S.-Y., K.-Y. Chwa, and S. Y. Shin. Image metamorphosis using snakes and free-form deformations. In: 22nd Annual Conference on Computer Graphics and Interactive Techniques. ACM, 1995.
Lee, S., G. Wolberg, and S. Y. Shin. Scattered data interpolation with multilevel B-splines. IEEE Trans. Visual Comput. Graphics 3:228–244, 1997.
Ma, J. J., A. Wittek, B. Zwick, G. R. Joldes, S. K. Warfield, and K. Miller. On the effects of model complexity in computing brain deformation for image-guided neurosurgery. In: Computational Biomechanics for Medicine: Soft Tissues and the Musculoskeletal System, edited by A. Wittek, P. M. F. Nielsen, and K. Miller. New York: Springer, 2011, pp. 51–61.
Mattes, D., D. R. Haynor, H. Vesselle, T. K. Lewellen, and W. Eubank. Nonrigid multimodality image registration. In: Medical Imaging: 2001: Image Processing, Pts 1–3, edited by M. Sonka and K. M. Hanson. pp. 1609–1620, 2001.
Mattes, D., D. R. Haynor, H. Vesselle, T. K. Lewellen, and W. Eubank. PET-CT image registration in the chest using free-form deformations. IEEE Trans. Med. Imaging 22:120–128, 2003.
Miller, K. Introduction. In: Biomechanics of the Brain, edited by K. Miller. New York: Springer, 2011, pp. 1–3.
Miller, K., and K. Chinzei. Mechanical properties of brain tissue in tension. J. Biomech. 35:483–490, 2002.
Miller, K., G. Joldes, D. Lance, and A. Wittek. Total Lagrangian explicit dynamics finite element algorithm for computing soft tissue deformation. Commun. Numer. Methods Eng. 23:121–134, 2007.
Miller, K., and J. Lu. On the prospect of patient-specific biomechanics without patient-specific properties of tissues. J. Mech. Behav. Biomed. 2013. doi:10/1016/j.jmbbm.2013.01.2013.
Miller, K., and A. Wittek. Neuroimage registration as displacement—zero traction problem of solid mechanics. In: Computational Biomechanics for Medicine, edited by K. Miller, and P. M. F. Nielsen. New York: Springer, 2006.
Nakaji, P., and R. F. Spetzler. Innovations in surgical approach: the marriage of technique, technology, and judgment. Clin. Neur. 51:177–185, 2004.
Rohlfing, T., C. R. Maurer, D. A. Bluemke, and M. A. Jacobs. Volume-preserving nonrigid registration of MR breast images using free-form deformation with an incompressibility constraint. IEEE Trans. Med. Imaging 22:730–741, 2003.
Rueckert, D., L. I. Sonoda, C. Hayes, D. L. G. Hill, M. O. Leach, and D. J. Hawkes. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging 18:712–721, 1999.
Ruprecht, D., and H. Muller. Free form deformation with scattered data interpolation methods. In: Geometric Modelling, edited by G. Farin, H. Hagen, H. Noltemeier, and W. Knodel. Berlin/Heidelberg: Springer-Verlag, 1993, pp. 267–281.
Schnabel, J., D. Rueckert, M. Quist, J. Blackall, A. Castellano-Smith, T. Hartkens, G. Penney, W. Hall, H. Liu, C. Truwit, F. Gerritsen, D. G. Hill, and D. Hawkes. A generic framework for non-rigid registration based on non-uniform multi-level free-form deformations. In: Medical Image Computing and Computer-Assisted Intervention, edited by W. Niessen, and M. Viergever. Berlin/Heidelberg: Springer, 2001, pp. 573–581.
Sinkus, R., M. Tanter, T. Xydeas, S. Catheline, J. Bercoff, and M. Fink. Viscoelastic shear properties of in vivo breast lesions measured by MR elastography. Magn. Reson. Imaging 23:159–165, 2005.
Thevenaz, P. and M. A. Unser. Spline pyramids for intermodal image registration using mutual information. In: Proceedings of SPIE: Wavelet Applications in Signal and Image Processing V. pp. 236–247, 1997.
Tokuda, J., G. S. Fischer, X. Papademetris, Z. Yaniv, L. Ibanez, P. Cheng, H. Liu, J. Blevins, J. Arata, A. J. Golby, T. Kapur, S. Pieper, E. C. Burdette, G. Fichtinger, C. M. Tempany, and N. Hata. OpenIGTLink: an open network protocol for image-guided therapy environment. Int. J. Med. Robotics Comput. Assist. Surg. 5:423–434, 2009.
Tustison, N. J., B. B. Avants, P. A. Cook, Y. Zheng, A. Egan, P. A. Yushkevich, and J. C. Gee. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29:1310–1320, 2010.
Ungi, T., P. Abolmaesumi, R. Jalal, M. Welch, I. Ayukawa, S. Nagpal, A. Lasso, M. Jaeger, D. P. Borschneck, G. Fichtinger, and P. Mousavi. Spinal needle navigation by tracked ultrasound snapshots. IEEE Trans. Biomed. Eng. 59:2766–2772, 2012.
Ungi, T., D. Sargent, E. Moult, A. Lasso, C. Pinter, R. C. McGraw, and G. Fichtinger. Perk Tutor: an open-source training platform for ultrasound-guided needle insertions. IEEE Trans. Biomed. Eng. 59:3475–3481, 2012.
Warfield, S. K., S. J. Haker, I. F. Talos, C. A. Kemper, N. Weisenfeld, A. U. J. Mewes, D. Goldberg-Zimring, K. H. Zou, C. F. Westin, W. M. Wells, C. M. C. Tempany, A. Golby, P. M. Black, F. A. Jolesz, and R. Kikinis. Capturing intraoperative deformations: research experience at Brigham and Women’s Hospital. Med. Image Anal. 9:145–162, 2005.
Wittek, A., T. Hawkins, and K. Miller. On the unimportance of constitutive models in computing brain deformation for image-guided surgery. Biomech. Model. Mechanobiol. 8:77–84, 2009.
Wittek, A., G. Joldes, M. Couton, S. K. Warfield, and K. Miller. Patient-specific non-linear finite element modelling for predicting soft organ deformation in real-time; Application to non-rigid neuroimage registration. Prog. Biophys. Mol. Biol. 103:292–303, 2010.
Wittek, A., K. Miller, R. Kikinis, and S. K. Warfield. Patient-specific model of brain deformation: application to medical image registration. J. Biomech. 40:919–929, 2007.
Wittek, A., and K. Omori. Parametric study of effects of brain-skull boundary conditions and brain material properties on responses of simplified finite element brain model under angular acceleration impulse in sagittal plane. JSME Int. J. 46:1388–1399, 2003.
Zhang, J. Y., G. R. Joldes, A. Wittek, and K. Miller. Patient-specific computational biomechanics of the brain without segmentation and meshing. Int. J. Numer. Methods Biomed. Eng. 29:293–308, 2013.
Zhao, C. J., W. K. Shi, and Y. Deng. A new Hausdorff distance for image matching. Pattern Recogn. Lett. 26:581–586, 2005.
Acknowledgments
The first author is a recipient of the SIRF scholarship and acknowledges the financial support of the University of Western Australia. The financial support of National Health and Medical Research Council (Grant No. APP1006031) is gratefully acknowledged. This investigation was also supported in part by NIH grants R01 EB008015 and R01 LM010033, and by a research grant from the Children’s Hospital Boston Translational Research Program. In addition, the authors also gratefully acknowledge the financial support of Neuroimage Analysis Center (NIH P41 EB015902), National Center for Image Guided Therapy (NIH U41RR019703) and the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from http://nihroadmap.nih.gov/bioinformatics.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Joel D. Stitzel oversaw the review of this article.
Rights and permissions
About this article
Cite this article
Mostayed, A., Garlapati, R.R., Joldes, G.R. et al. Biomechanical Model as a Registration Tool for Image-Guided Neurosurgery: Evaluation Against BSpline Registration. Ann Biomed Eng 41, 2409–2425 (2013). https://doi.org/10.1007/s10439-013-0838-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-013-0838-y