Abstract
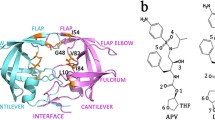
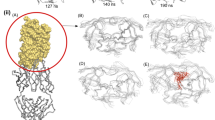
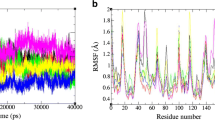
The emergence of drug-resistant mutants of HIV-1 is a tragic effect associated with conventional long-treatment therapies against acquired immunodeficiency syndrome. These mutations frequently involve the aspartic protease encoded by the virus; knowledge of the molecular mechanisms underlying the conformational changes of HIV-1 protease mutants may be useful in developing more effective and longer lasting treatment regimes. The flap regions of the protease are the target of a particular type of mutations occurring far from the active site. These mutations modify the affinity for both substrate and ligands, thus conferring resistance. In this work, molecular dynamics simulations were performed on a native wild type HIV-1 protease and on the drug-resistant M46I/G51D double mutant. The simulation was carried out for a time of 3.5 ns using the GROMOS96 force field, with implementation of the SPC216 explicit solvation model. The results show that the flaps may exist in an ensemble of conformations between a “closed” and an “open” conformation. The behaviour of the flap tips during simulations is different between the native enzyme and the mutant. The mutation pattern leads to stabilization of the flaps in a semi-open configuration.





Similar content being viewed by others
References
Seelmeier S, Schmidt H, Turk V, Von der Helm K (1988) Proc Natl Acad Sci USA 85:6612–6616
Huff JR (1991) J Med Chem 34:2305–2314
Tozzini V, Trylska J, Chang C, McCammon JA (2007) J Struct Biol 157:606–615
Chatfield DC, Brooks BR (1995) J Am Chem Soc 117:5561–5572
Silva AM, Cachau RE, Sham HL, Erickson JW (1996) J Mol Biol 255:321–340
Chatfield DC, Eurenius KP, Brooks BR (1998) J Mol Struct 423:79–92
Ohtaka H, Schon A, Freire E (2003) Biochemistry 42:13659–13666
Scott WRP, Schiffer CA (2000) Structure 8:1259–1265
Spinelli S, Liu QZ, Alzari PM, Hirel PH, Poljak RJ (1991) Biochimie 73:1391–1393
Oostenbrink C, Villa A, Mark AE, Van Gusteren WF (2004) J Comput Chem 25:1656–1676
Berendsen HJC, Van der Spoel D, Van Drunen R (1995) Comput Phys Commun 91:43–56
Lindahl E, Hess B, Van der Spoel D (2001) J Mol Model 7:306–317
Berendsen HJC, Postma JPM, DiNola A, Haak JR (1984) J Chem Phys 81:3684–3690
Ferguson DM (1995) J Comp Chem 16:501–511
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) J Chem Phys 103:8577–8593
Pedretti A, Villa L, Vistoli, G (2002) J Mol Graph 21:47–49
Humphrey W, Dalke A, Schulten K (1996) J Mol Graph 14:33–38
Perryman AL, Lin J-H, McCammon A (2004) Protein Sci 13:1108–1123
Meiselbach H, Horn AHC, Harrer T, Sticht H (2007) J Mol Model 13:297–304
Maschera B, Darby G, Palu G, Wright LL, Tisdale M, Myers R, Blair ED, Fufine ES (1996) J Biol Chem 271:33231–33235
Piana S, Carloni P, Rothlingsberger U (2002) Protein Sci 11:2393–2402
Wu TD, Schiffer CA, Gonzales MJ, Taylor J, Kantor R, Chou S, Israeliski D, Zolopa AR, Fessel WJ, Shafer RW (2003) J Virol 77:4836–4847
Tòth G, Borics A (2006) J Mol Graph Model 24:465–474
Ingr M, Uhlìkovà T, Strisovsky K, Majerovà E, Konvalinka J (2003) Protein Sci 12:2173–2182
Hornak V, Okur A, Rizzo RC, Simmerling C (2006) Proc Natl Acad Sci USA 103:915–920
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lauria, A., Ippolito, M. & Almerico, A.M. Molecular dynamics studies on HIV-1 protease: a comparison of the flap motions between wild type protease and the M46I/G51D double mutant. J Mol Model 13, 1151–1156 (2007). https://doi.org/10.1007/s00894-007-0242-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-007-0242-3