Abstract
The integrated stress response (ISR), a defense mechanism cells employ when under stress (e.g., amino acid deprivation), causes suppression of global protein synthesis along with the paradoxical increased expression of a host of proteins that are useful in combating various stresses. Genes that were similarly differentially expressed under conditions of either leucine- or cysteine-depletion were identified. Many of the genes known to contain an amino acid response element and to be induced in response to eIF2α phosphorylation and ATF4 heterodimer binding (ATF3, C/EBPβ, SLC7A1, SLC7A11, and TRIB3), as well as others shown to be induced downstream of eIF2α phosphorylation (C/EBPγ, CARS, SARS, CLCN3, CBX4, and PPP1R15A) were among the upregulated genes. Evidence for the induction of the ISR in these cells also included the increased phosphorylation of eIF2α and increased protein abundance of ATF4, ATF3, and ASNS in cysteine- and leucine-depleted cells. Based on genes highly differentially expressed in both leucine- and cysteine-deficient cells, a list of 67 downregulated and 53 upregulated genes is suggested as likely targets of essential amino acid deprivation in mammalian cells.
Similar content being viewed by others
Introduction
A fundamental question in biology is how cells sense and respond to changes in their nutrient supply. How mammalian cells respond to changes in the availability of essential amino acids has been a major research focus in recent years due to evidence for various roles of essential amino acids in the regulation of protein translation and growth as well as regulation of stress response pathways.
In our studies of cysteine metabolism in mammalian cells, we discovered that cysteine deprivation does not necessarily induce an oxidative state in unstressed cells (Lee et al. 2008) but, rather, the data suggested that expression of genes for cyst(e)ine uptake and GSH synthesis may be induced as part of the cell’s normal response to amino acid deficiency. In particular, we observed highly significant upregulation of expression of a group of genes (ASNS, ATF3, C/EBPβ, SLC7A11, TRIB3, ATF4, and SLC7A1) that are known to contain an amino acid response element (AARE) and to respond to amino acid deprivation via the binding of activating transcription factor 4 (ATF4).
ATF4 is translationally, as well as transcriptionally, upregulated in response to activation of stress response eIF2α kinases which then phosphorylate Ser51 of the alpha subunit of eukaryotic initiation factor 2 (eIF2). One of the four mammalian eIF2α kinases is GCN2 (protein kinase general control nonderepressible 2, or eIF2α kinase 4) which is activated specifically in response to amino acid deprivation (Wek et al. 2006). The GCN2 kinase senses an increase in the abundance of non-aminoacylated tRNAs via its histidyl-tRNA synthetase homologous domain (Wek et al. 1995; Padyana et al. 2005; Hao et al. 2005). Binding of uncharged tRNAs to this domain activates the juxtaposed protein kinase domain of GCN2, rendering the kinase active and leading to phosphorylation of Ser51 of eIF2α. As shown in Fig. 1a, the phosphorylation of eIF2 subsequently leads to inhibition of the GTP-GDP exchange activity of eIF2B, and this in turn leads to diminished formation of the ternary complex (Met-tRNA Meti ·eIF2 –GTP) that is needed for all cytoplasmic translation initiation events (Gabauer and Hentze 2004; Hinnebusch 2000; Kubica et al. 2006; Kimball et al. 1991). In addition to globally reducing translation initiation events and hence the overall rate of protein synthesis, reduced ternary complex formation paradoxically results in increases in the translation of some specific mRNAs, most notably that for ATF4 (Dang Do et al. 2009), as illustrated in Fig. 1b. The presence of an inhibitory upstream open reading frame (ORF) that overlaps the ATF4 ORF promotes read-through of the ATF4 translational start codon, preventing ATF4 translation under normal conditions in which ternary complex is abundant (Lu et al. 2004a, b; Wek et al. 2006). In amino acid deprivation, in which eIF2B is inhibited and ternary complex formation is reduced, ribosomal scanning through the inhibitory upstream ORF start codon is favored so that initiation occurs at the downstream ATF4 ORF start codon, allowing an increase in ATF4 mRNA translation.
Mechanisms by which eIF2α kinases downregulate formation of ternary complex (eIF2-GDP·Met-tRNA Meti ) formation (a) and by which the reduced availability of ternary complex suppresses global protein translation while selectively increasing ATF4 mRNA translation (b). The increase in ATF4 translation leads to induction of genes containing amino acid response elements (AARE). The collective consequences of eIF2α phosphorylation are termed the integrated stress response (ISR)
This translationally mediated increase in ATF4 leads to a transcriptional response that involves the upregulation of a number of genes including that for ATF4 itself (Lu et al. 2004a, b; Harding et al. 2000, 2003). GCN2 is the only eIF2α kinase in yeast, and activation of GCN2 in yeast results in increased translation of GCN4, the functional counterpart to mammalian ATF4, and GCN4 acts to induce expression of an array of genes that encode amino acid biosynthetic enzymes (Hinnebusch 2005). Unlike yeast, mammalian cells express several different eIF2α kinases, which are activated by various kinds of stress including accumulation of unfolded proteins, heme deficiency, and viral infection, in addition to amino acid deprivation (Wek et al. 2006). Because mammalian cells, unlike yeast, are unable to synthesize a subset of amino acids, the so-called essential or indispensable amino acids, they must obtain them from the environment. Not surprisingly, the phospho-eIF2α/ATF4 signaling cascade induces the expression of a somewhat different array of genes in mammalian cells compared with those induced by the GCN2/GCN4 pathway in yeast. The genes induced in mammalian cells include those encoding transcriptional regulators, solute carrier family transporters, aminoacyl-tRNA synthetases, and proteins involved in cell cycle and DNA repair (Harding et al. 2000, 2003; Lee et al. 2008; Lu et al. 2004b; Sato et al. 2004). Because the mammalian response to eIF2α phosphorylation is induced by kinases that respond to a variety of stressors, the downstream translational/transcriptional response is often termed the integrated stress response (ISR).
Although amino acid deprivation and other stresses that activate mammalian eIF2α kinases commonly trigger increased ATF4 synthesis, a somewhat distinct subset of genes is activated in response to different types of cellular stress, perhaps due to eIF2α kinase-independent signaling, which, among other things, may lead to induction of different C/EBP family members (Wek et al. 1995; Dang Do et al. 2009; Shan et al. 2009). Gene expression surveys have been conducted with cells lacking ATF4 (Harding et al. 2003), in cells with ER-stress induced by tunicamycin (Harding et al. 2003; Lu et al. 2004b), with drug-induced activity of an artificial eIF2α kinase, Fv2E-PERK (Lu et al. 2004b), and in cells lacking GCN2 (Deval et al. 2009). All of these studies have been done using immortalized murine fibroblasts. To our knowledge, no large gene expression surveys have been reported for mammalian cells exposed to amino acid deprivation other than our studies in cysteine-deficient HepG2/C3A cells (Lee et al. 2008). Results could vary with both the type of stress and with the type of cell that is used for the gene expression studies.
Thus, to further assess the role of amino acid deprivation on eIF2α phosphorylation and downstream transcriptional responses, we conducted additional microarray studies for leucine-depleted HepG2/C3A cells and compared these with results for cysteine-depleted cells. The comparison of cells exposed to deficiencies of two different amino acids was done to facilitate selection of genes whose expression is more likely to be altered specifically in response to GCN2-induced eIF2α phosphorylation. In addition, we assessed the effects of both cysteine and leucine deprivation on the phosphorylation of eIF2α and on the protein expression patterns of ATF4 and other ISR-related proteins. The results of this study add to our understanding of how gene expression changes in response to amino acid deprivation in a human-derived cell line. Amino acid deprivation frequently induces a milder degree of stress than the agents used to induce ER stress (e.g., tunicamycin, thapsigargin), and the human HepG2/C3A cell line represents a more-differentiated cell line than the immortalized murine embryonic fibroblasts used in the studies mentioned above. The HepG2/C3A cell line is a derivative of the human hepatocarcinoma HepG2 cell line, with the C3A derivative being selected for its more liver-specific phenotype. HepG2 cells have been used in other studies of the amino acid deprivation response (Jousse et al. 2000; Thiaville et al. 2008; Palii et al. 2009).
Materials and methods
Experimental treatment of HepG2/C3A cells
HepG2/C3A cells (ATCC CRL-10741) were cultured in a humidified incubator at 37°C and 5% CO2 in complete medium prepared using sulfur amino acid (SAA)- and leucine-free Dulbecco’s modified Eagle’s medium (DMEM; from Gibco/Invitrogen) supplemented with 0.8 mM l-leucine, 0.2 mM l-methionine, 0.2 mM l-cysteine, 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 1× MEM nonessential amino acid solution and 0.05 mM bathocuproine disulfonate. All cells were plated in complete medium at a density of 1 × 106 cells per 100 mm diameter culture dish. After 24 h of culture in complete medium to allow the cells to reach 50–60% confluence, the medium was replaced with experimental medium, which was the same complete medium (+Leu, +Cys) or medium prepared without leucine (−Leu) or without cysteine (−Cys). Cells were cultured with a medium change at 18 h and harvested at the indicated time-points.
RNA extraction, microarray analysis, and selection of differentially expressed genes
HepG2/C3A cells cultured in complete or Leu-free treatment medium for 36 h as described above were washed twice with ice-cold PBS and then directly lysed into denaturation solution. Total RNA was extracted from three separate plates of cells grown under each treatment with an RNeasy Micro kit (Qiagen). Microarray analysis was performed on each of the three samples for each treatment using Affymetrix GeneChip Human plus 2.0 Arrays, a GeneChip Scanner 3000, and Affymetrix GCOS software, as described previously (Lee et al. 2008). To look for differential gene expression between the two treatment conditions, the Student’s t test was applied on the normalized signals for each probe set. To select the differential gene set, a significance level cutoff was empirically established at P < 0.0001, based on the number of genes passing the cutoff, the magnitude of fold changes, and the relationship between fold changes and P values. The complete set of microarray data has been deposited in the NCBI Gene Expression Omnibus (accession no. GSE13142, http://www.ncbi.nlm.nih.gov/geo/).
Comparison of gene expression array results for HepG2/C3A cells cultured in leucine-free medium versus complete medium with those for HepG2/C3A cells cultured in cysteine-free medium versus complete medium
Using a dataset generated earlier for HepG2/C3A cells cultured in cysteine-free (−Cys) medium (SAA-free DMEM supplemented with 0.1 mM l-methionine) versus cells cultured in cysteine-supplemented (+Cys) medium (SAA-free DMEM supplemented with 0.1 mM l-methionine plus 0.1 mM l-cysteine) (GEO, accession no. GSE9517), we similarly selected differentially expressed genes (P ≤ 0.0001). The two sets of selected genes were then compared to each other to select those that were differentially expressed (P ≤ 0.0001) in both Cys-deficient and Leu-deficient HepG2/C3A cells. It should be noted that HepG2/C3A cells are readily depleted of cysteine, even when cultured in methionine-containing medium, due to the lack of the high K m isoform of methionine adenosyltransferase (Lee et al. 2008; Lu and Huang 1994).
Western blot analysis of protein levels in HepG2/C3A cells cultured in −Leu, −Cys, or complete medium
Dishes of HepG2/C3A cells were cultured in complete, −Leu, or −Cys medium as described earlier. After 6, 12, 24, or 30 h of culture, treatment medium was aspirated and cells were washed with ice-cold PBS containing 10 mM NaF. Monolayers were then harvested into TNESV lysis solution (50 mM Tris, pH 7.5, 1% (v/v) Nonidet P-40, 2 mM EDTA, 150 mM NaCl, and 10 mM sodium orthovanadate) supplemented with 1× PhosSTOP phosphatase inhibitor cocktail (Roche Applied Science) and 1× Complete Protease Inhibitor Cocktail (Roche). Cell lysates were centrifuged at 17,000×g for 30 min, and the protein concentration of the supernatants was determined using the bicinchonic acid assay (Pierce). For western blotting, 60 μg of total supernatant protein from each sample were separated by one-dimensional SDS-PAGE (12% w/v acrylamide) and electroblotted overnight onto 0.45 μm (pore size) Immobilon-P PVDF membranes (Millipore). Membranes were immunoblotted for proteins of interest using the following antibodies: anti-pS51-eIF2α and anti-eIF2α (total) from Cell Signaling Technology; anti-ATF4 (gift from M.S. Kilberg, University of Florida, Gainesville, FL); anti-ATF3 (C-19), anti-ASNS (C-14), anti-CHOP, and anti-HSP5A from Santa Cruz Biotechnology; and anti-GCLC from Neomarkers (Freemont, CA). Bands were visualized using horseradish peroxidase-coupled secondary antibodies and chemiluminescent substrates (West Dura, Pierce) and autoradiography. The cell culture studies were repeated three or more times for designated time-points to assure the repeatability of the results.
Results
Effect of amino acid deficiency on gene expression in HepG2/C3A cells
A total of 670 genes were identified as differentially expressed with a P ≤ 0.0001 in cells cultured in leucine deficient (−Leu) versus complete (+Leu) medium, with 301 being upregulated and 369 being downregulated (Tables S1 and S2 in supplemental materials). Similarly, 807 genes were identified as differentially expressed with a P ≤ 0.0001 in cells cultured in cysteine deficient (−Cys) versus complete (+Cys) medium (GSE13142 data set; Tables S3 and S4 in supplemental materials); 407 genes were upregulated and 400 were downregulated. To obtain a list of genes that are potential targets of regulated expression in response to essential amino acid deprivation, genes that met the following conditions were selected: (1) the gene was differentially expressed at P ≤ 0.0001 in both the −Leu and the −Cys conditions; (2) the differential expression of the gene was at least twofold; and (3) the differential expression of the gene was detected with the same probe set in both series of studies. Application of these criteria yielded the list of 120 genes that are listed in Table 1. Of these genes that were differentially expressed in response to both leucine and cysteine deprivation, 67 were downregulated. This downregulated group includes many genes involved in cell cycle/cell division, fatty acid and sterol metabolism/synthesis, and nucleotide metabolism/synthesis, as shown in Table 1. The remaining 53 genes were upregulated, and this upregulated group of genes includes a number of genes involved in amino acid uptake, aminoacyl-tRNA synthesis, transcriptional regulation, and growth inhibition. Most notably, many of the genes known to contain AAREs and to be induced in response to eIF2α (Ser51) phosphorylation and ATF4 heterodimer binding (i.e., ATF3, C/EBPβ, SLC7A1, SLC7A11, and TRIB3), as well as others shown to be induced downstream of eIF2α (Ser51) phosphorylation (C/EBPγ, CARS, SARS, CLCN3, CBX4, and PPP1R15A, which is also known as GADD34, were among these 53 genes (Lee et al. 2008; Lu et al. 2004b) (see Table 1 for names of gene products). Expression of ASNS, another AARE-dependent gene, met all criteria except one in that the fold increase for expression in the −Cys medium was only 1.6. Two other well-characterized AARE-containing genes, ATF4 and (also known as DDIT3 or GADD153) were also identified as significantly differentially expressed in cells cultured in both −Leu and −Cys medium using a lower statistical significance cutoff of P ≤0.001.
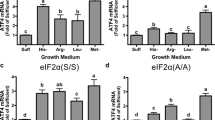
Confirmation that the amino acid deprivation response was actually initiated in HepG2/C3A cells cultured in −Leu or −Cys medium was obtained by western blot analysis of total and phosphorylated eIF2α, ATF4, activating transcription factor 3 (ATF3), and asparagine synthase (ASNS) proteins in the cells. Glutamate-cysteine ligase catalytic subunit (GCLC) and heat shock 70-kDa protein 5 (HSPA5) were also assayed to confirm that proteins that are not part of the amino acid deprivation response were not upregulated. As shown in Fig. 2, the amount of phosphorylated eIF2α increased between 6 and 24 h of culture in cells cultured in −Leu or −Cys, as well as in control medium. However, the increases were greater in cells cultured in −Leu or −Cys medium than in cells cultured in control medium, such that the amount of phosphorylated eIF2α in cells cultured for 24 to 30 h in amino-acid-deficient medium was double that in cells cultured in control medium (2.0-times for −Leu and 2.2-times for −Cys). The amount of total eIF2α did not change significantly across time or culture conditions. A significant increase in total ATF4 levels in the cells was noted only at the 24 h time-point at which time ATF4 in cells cultured in −Leu medium was 1.3-times and that in cells cultured in −Cys medium was 1.2-times that in control cells. Expression of ATF3 and ASNS, whose genes are well-established targets for upregulation in response to amino acid deprivation or eIF2α phosphorylation, paralleled the increases in eIF2α phosphorylation. ATF3 levels in cells cultured in amino acid-deficient medium for 24 h/30 h were 8.0- and 9.8-times for −Leu and −Cys cells, respectively, and ASNS levels were 2.7 and 1.9-times, respectively, those in cells cultured in complete medium. CHOP was strongly induced in cells cultured in −Leu medium but only weakly detected in cells cultured in −Cys medium at the longest time-point (30 h). The amount of GCLC, which served as a loading control, did not change with time or treatment. As a control on the specificity of the amino acid deprivation/GCN2-mediated response, a chaperone protein that is strongly induced in response to activation of a different eIF2α kinase (i.e., PERK, or eIF2α kinase 3) by a different type of stress (endoplasmic reticulum stress, or ER stress) was measured. The ER chaperone HSPA5 was not increased over time or with amino acid deprivation of the medium and, in fact, was consistently lower in cells cultured in −Leu medium than in those cultured in control medium. Similar results were obtained for HepG2/C3A cells cultured in medium deficient in l-methionine (but containing 0.2 mM l-cysteine) (data not shown).
Comparison of results for genes induced in response to amino acid deficiency with lists of upregulated genes previously generated for eIF2α phosphorylation/ATF4-mediated changes in gene expression
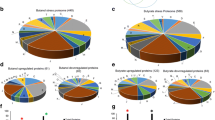
To further explore the list of 53 upregulated genes that met stringent criteria for designation as differentially expressed (P ≤ 0.0001) in HepG2/C3A cells cultured in amino acid deficient medium, this list of genes was compared with lists of upregulated genes previously generated for eIF2α phosphorylation/ATF4-mediated changes in gene expression (Fig. 3). These comparison lists include (1) that for murine fibroblasts that was generated by Harding et al. (2003) by selecting genes whose expression was at least twofold lower in tunicamycin-treated ATF4−/− cells compared with tunicamycin-treated wild-type cells (i.e., genes upregulated by tunicamycin in an ATF4-dependent fashion) and (2) that for murine fibroblasts that was generated by Lu et al. (2004b) by selecting genes whose expression was induced at least twofold in both drug-induced Fv2E-PERK cells and 1.5-fold in tunicamycin-treated wild-type cells (i.e., genes upregulated by both drug-induced eIF2α phosphorylation and by induction of ER stress). Interestingly, only two genes appear on all three lists: C/EBPβ and C/EBPγ (CCAAT/enhancer binding protein, β and γ, respectively). These two genes represent the only overlap with the selected list of Harding et al. (2003). Comparison of our list with that of Lu et al. (2004b) yields seven additional overlapping genes: ATF3, SARS (seryl-tRNA synthetase), CARS (cysteinyl-tRNA synthetase), CBX4 (chromobox homolog 4), CLCN3 (chloride channel 3), LMO4 (LIM domain only 4), and TXNIP (thioredoxin interacting protein). ASNS (asparagine synthetase), SLC1A7 (glutamate/neutral amino acid transporter), NARS (asparaginyl-tRNA synthetase), and ERO1L (endoplasmic reticulum 1-like oxidoreductase) made the lists of both Harding et al. (2003) and Lu et al. (2004a, b) but not our list of genes induced by amino acid deprivation in HepG2/C3A cells.
Comparison of genes proposed to be induced in response to eIF2α phosphorylation and/or ATF4 induction. Lists of upregulated genes generated by Harding et al. (2003) by selecting genes whose expression was at least twofold lower in tunicamycin-treated ATF4−/− murine fibroblasts compared with tunicamycin-treated wild-type cells; Lu et al. (2004b) by selecting genes whose expression was induced at least twofold in both drug-induced Fv2E-PERK murine fibroblasts and 1.5-fold in tunicamycin-treated wild-type cells; and in our work by selecting genes that were induced at least twofold in HepG2/C3A cells when cultured in either cysteine-deficient medium or leucine-deficient medium
Discussion
Amino acid deprivation leads to downregulation of genes involved in amino acid uptake, aminoacyl-tRNA synthesis, transcriptional regulation, and growth inhibition
The 120 genes that met stringent criteria for designation as differentially expressed (P ≤ 0.0001) in HepG2/C3A cells cultured in amino acid deficient medium should all be considered potential targets for regulation by amino acids in HepG2/C3A cells.
The upregulated group of 53 genes includes a number of genes involved in amino acid uptake, aminoacyl-tRNA synthesis, transcriptional regulation, and growth inhibition; comparison of this group of upregulated genes with the groups of upregulated genes identified by other investigators as potential downstream targets of eIF2α phosphorylation and ATF4 induction, amino acyl-tRNA synthetases and amino acid transporters stand out as consistently upregulated although the specific ones varied among studies. The cystine-glutamate exchanger, SLC7A11, was the most highly induced amino acid transporter gene in HepG2/C3A cells in both −Cys and −Leu medium. Several amino acyl-tRNA synthetases, CARS, SARS, and WARS, were induced under conditions of both Cys and Leu deficiency.
Another striking observation from our comparison of various studies to identify common genes induced by various stresses that induce eIF2α phosphorylation and ATF4 expression is that a number of genes that are known to contain AAREs and to be induced by ATF4 binding were identified, as would be anticipated. These include C/EBPβ, ATF3, and ASNS. Other genes induced in the amino acid deficient HepG2/C3A cells that are known to contain AAREs and to be induced by ATF4 binding include SLC7A1, SLC7A11, and TRIB3. The identification of these AARE-containing genes is consistent with transcriptional responses to amino acid deprivation being mediated by ATF4 binding to AARE and with phospho-eIF2α/ATF4-dependent gene expression being a key component of the ISR.
Further support for the induction of the ISR in HepG2/C3A cells cultured in amino acid deficient medium was obtained from the eIF2α phosphorylation and protein expression patterns in the HepG2/C3A cells. Increases in eIF2α phosphorylation were apparent by 12 h after the removal of leucine or cysteine from the medium as were increases in ATF3 protein levels, and increases in ATF4 and ASNS protein levels were apparent in both cysteine- and leucine-depleted cells by 24 h. Whereas ATF3 protein was strongly induced by either leucine deprivation or cysteine deprivation, ASNS protein was consistently more strongly induced in the cysteine-depleted cells. On the other hand, CHOP protein was strongly induced in the leucine-depleted cells whereas induction was not evident in the cysteine-depleted cells until the 30-h time-point. The stronger induction of CHOP by leucine depletion is consistent with our microarray results that showed a 6.7-fold induction (significant at P ≤ 0.0001) of CHOP mRNA in −Leu-treated cells but only a 4.1-fold, less significant (P ≤ 0.001) induction in −Cys-treated cells. Jousse et al. (2000) previously reported a similar finding for both HeLa and HepG2 cells, finding that CHOP protein induction was greater in cells incubated for 16 h in −Leu medium than in cells incubated in −Cys medium. Thus, a robust upregulation of components of the amino acid deprivation pathway or ISR was observed, although not all downstream responses were highly correlated with the degree of eIF2α phosphorylation or with cellular ATF4 levels.
There are several possible reasons for the lack of apparent correlation of all upstream responses with activation or expression of early components of the ISR pathway or of mRNA and protein expression levels for individual genes. First of all, the time-course for changes in levels of various mRNAs and proteins is highly dependent upon the turnover rate of the individual mRNAs and proteins. Second, the levels of ATF4 in the nucleus may not parallel the amount of total ATF4 in the cell. Third, other transcriptional regulators including ATF4’s heterodimerization partners may also play an important role in gene activation. ATF4 forms heterodimers with members of another bZIP family, which consists of C/EBP α, β, γ, δ, ε, and CHOP. ATF4-C/EBP complexes have been detected at the C/EBP-ATF composite sites (functional AAREs) on promoters of genes induced by amino acid deprivation. ATF4 activity is subject to regulation (activation or repression) by its various binding partners, with some of these dimerization partners (e.g., ATF3, TRIB3, C/EBPβ, and CHOP), as well as the ATF4 gene itself, being transcriptionally induced by ATF4 heterodimers (Shan et al. 2009; Mungrue et al. 2009; Pan et al. 2007). Fourth, some of the effects of amino acid deprivation are undoubtedly mediated by GCN2-independent pathways and these may vary for different amino acids (Jousse et al. 2000; Su et al. 2009; Deval et al. 2009; Thiaville et al. 2008; Palii et al. 2009). Finally, regulation of gene expression and/or protein level may involve the integration of a number of regulators and points of regulation, with these additional signaling pathways either enhancing or suppressing the GCN2/phospho-eIF2α/ATF4 signaling pathway (Shan et al. 2009; Ishikawa et al. 2009; Vesely et al. 2009).
Amino acid deprivation leads to downregulation of genes involved in the cell cycle, fatty acid desaturation, sterol synthesis, and nucleotide synthesis
The group of genes that were downregulated in cells cultured in either −Leu or −Cys medium included a number of genes involved in cell cycle/cell division, fatty acid and sterol metabolism/synthesis, and nucleotide metabolism/synthesis (Table 1). Genes that are downregulated in response to eIF2α phosphorylation/ATF4 induction have generally received much less attention, although one would expect that DNA synthesis and cell proliferation and growth would be restricted under conditions of cell stress so that resources can be diverted to deal with the imposed stress. Downregulation of the expression of genes involved in cell division and nucleotide synthesis, along with the eIF2α phospho-mediated reduction in protein synthesis, would seem to be synergistic in facilitating the cell’s survival during times of nutrient deprivation or other stress. We observed downregulation of expression of a cluster of genes in the folate/deoxythymidine nucleotide synthetic pathway as well as downregulation of genes involved in all phases of the cell cycle.
Guo and Cavener (2007) speculated that GCN2 may act as a master regulator of metabolic adaptation to nutrient deprivation, because an essential amino acid limitation in nature would almost always be correlated with the deprivation of other nutrients as well. Using GCN2−/− and wild-type mice, they found that triglyceride synthesis was repressed in the livers of wild-type mice that were fed a leucine-devoid diet for 7 or 17 days but continued unabated in the livers of GCN2−/− mice. In the GCN2−/− mice, consumption of a leucine-deficient diet was accompanied by accumulation of hepatic triglyceride as well as impaired lipolysis of adipose stores. Guo and Cavener (2007), using quantitative RT-PCR and western blotting, showed that the −Leu diet resulted in diminished fatty acid synthase (FAS) expression in wild-type but not GCN2−/− mice. Similarly, hepatic mRNA levels for stearoyl-CoA desaturase (SCD) and other enzymes involved in fatty acid synthesis were diminished in GCN2−/− mice fed the −Leu diet but remained the same or even increased in wild-type mice. Furthermore, they showed that the mRNA levels for enzymes involved in β-oxidation (e.g., fatty acyl-CoA oxidase, long- and medium-chain acyl-CoA dehydrogenases) were not affected by leucine deprivation in wild-type mice, whereas the expression of these genes was markedly increased in GCN2−/− mice fed a leucine-devoid diet. Our gene expression studies in HepG2/C3A cells cultured in medium deficient in either Cys or Leu also showed the consistent downregulation of a group of genes involved in fatty acid and sterol metabolism, although the particular genes showing high fold induction in our microarray study were somewhat different than those observed by Guo and Cavener. The genes whose mRNA levels were significantly reduced (P ≤ 0.0001, ≤0.5-fold) in our analysis included genes involved in fatty acid desaturation (FADS1 and SCD), in isoprenoid and sterol synthesis (FDFT1, IDI1, LSS, EBP, SQLE, and SC4MOL), and in cholesterol ester synthesis (ACAT2). The only gene in this repressed group that was also included in the more-limited screen done by Guo and Cavener (2007) is SCD, and they found that SCD mRNA level was reduced to <20% of control in liver of wild-type mice fed the leucine-devoid diet. Furthermore, Guo and Cavener (2007) showed that this reduction was strictly dependent on GCN2 because SCD mRNA levels were not different in GCN2−/− mice fed the leucine-devoid versus control diet. Guo and Cavener (2007) found that expression of SREBP-1c mRNA and protein was repressed by leucine deprivation in liver of wild-type mice but not in GCN2−/− mice, suggesting that regulation of SREBP-1c may underlie some of the other transcriptional changes in genes involved in fatty acid synthesis. They did not, however, pursue the role of SREBP-2 in the regulation of sterol synthesis in their study which focused of regulation of triglyceride synthesis. Nevertheless, our findings support their hypothesis that amino acid deficiency has effects on overall macronutrient metabolism and, in particular, leads to a restriction of lipid synthesis.
Deprivation of a single amino acid likely results in amino acid-specific changes in gene expression in addition to those mediated by the ISR
In the previously reported analysis of −Cys/+Cys microarray results, we selected genes that were differentially expressed under conditions of both 6- and 42-h culture in treatment medium. Of the 24 genes significantly differentially expressed (P ≤ 0.0001) after both 6- and 42-h culture in −Cys/+Cys medium (Table 2 of Lee et al. 2008), 16 remained on the list of genes differentially expressed in both the −Leu/+Leu and −Cys/+Cys conditions reported here. All of the genes known to contain functional AAREs that were differentially expressed in response to both short- and long-term culture in −Cys medium (ATF3, C/EBPβ, SLC7A11, SLC7A1, and TRIB3), along with CARS, MAP1LC3B, VLDLR, ARHGEF2, INHBE, DAF, CBX4. C/EBPγ, FLJ10618, MGC16635, and NPIP///LOC339047//LOC440341, were also significantly upregulated in response to leucine deprivation in this study. ASNS, FOXO3A, GADD45A, GCLM, LNK, STC2, TUBE1, TXNRD1, and ZFAND1, which were significantly upregulated in response to both 6- and 42-h culture in −Cys medium were not detected as significantly upregulated in response to −Leu. These genes might be more responsive to cysteine depletion than to depletion of leucine or possibly other amino acids. GLCM and TXNRD1 contain known EpREs and may have been responding to glutathione depletion caused by cysteine deprivation.
Identification of transcriptionally regulated targets of the phospho-eIF2α/ATF4 pathway
Comparison of the genes identified in this study with those previously identified in different studies also aimed at identification of genes that respond to eIF2α phosphorylation and ATF4 induction yielded a limited number of genes that were identified in more than one study (see Fig. 3). These include C/EBPβ, C/EBPγ, ATF3, SARS, CARS, CBX4, CLCN3, LMO4, and TXNIP, which were identified in this study, as well as ASNS, SLC1A7, NARS, and ERO1L, which were identified in two studies of murine fibroblasts but not in our study with HepG2/C3A cells. It is quite interesting that only three of these 13 genes (C/EBPβ, ATF3, and ASNS) have been extensively studied as ISR target genes. C/EBPγ, CBX4, and LMO4 may also be important transcriptional regulators of the ISR. Amino acyl-tRNA synthetases, including CARS and SARS, are likely targets and would serve to preserve synthesis of critical stress-response proteins in the face of amino acid deprivation. The roles of induction of CLCN3 and TXNIP expression are less clear, and confirmation that levels of these proteins increase is still needed. TXNIP binds reduced thioredoxin and inhibits the ability of thioredoxin to reduce other proteins, thus increasing redox stress, inhibiting growth and hypertrophy, and causing increased apoptosis (Patwari et al. 2006). CLCN3 has been localized to late endosomes/lysosomes where it apparently is involved in organelle acidification (Li et al. 2002; Okamoto et al. 2008), and this would be consistent with the increase in macroautophagy observed in nutrient-depleted cells (Tallóczy et al. 2002; Kuma et al. 2004; Komatsu et al. 2005).
Summary
The ISR is generally viewed to be adaptive and to promote cell survival in the face of stress. The downregulation of expression of genes involved in cell growth and division, as well as the global reduction in protein synthesis that is a consequence of eIF2α phosphorylation, is consistent with cell survival under conditions of nutrient limitation. At the same time, the induction of expression of genes involved in amino acid uptake and aminoacyl-tRNA synthesis and reduced expression of lipogenic genes suggest that eukaryotic cells may attempt to adapt to an amino acid- or nutrient-limited environment by facilitating more efficient utilization of the limited resources (i.e., amino acids, other nutrients) and upregulating stress response genes, while at the same time suppressing overall protein synthesis, cell proliferation, and growth. All of these responses can be seen as consistent with cell survival under conditions of nutrient limitation.
We have compiled a list of 120 genes whose expression was differentially expressed in HepG2/C3A cells cultured in either cysteine- or leucine-deficient medium, and this list contains many of the genes known to respond to eIF2α kinase activation and to be components of the ISR. Clearly, amino acid deficiency, including cysteine deficiency in the absence of oxidative stress, induces eIF2α phosphorylation, presumably by activation of GCN2, leading to changes in expression of ATF4 and stress-related target genes. This list of target genes should be useful, as an initial step, in identifying potential components of the ISR as well as possibly identifying some genes that are differentially expressed in response to amino acid deprivation but not other stress conditions. Comparison of genes that were highly upregulated in HepG2/C3A cells cultured in −Cys and also in −Leu medium with lists generated by other investigators for genes induced in murine fibroblasts in response to eIF2α phosphorylation yielded a small list of only nine common genes (C/EBPβ, C/EBPγ, ATF3, SARS, CARS, CBX4, CLCN3, LMO4, and TXNIP). These nine genes are likely to be robust targets of the phospho-eIF2α/ATF4-mediated pathway of transcriptional regulation, although only two of them (C/EBPβ and ATF3) have so far been well characterized as target genes.
Abbreviations
- ISR:
-
Integrated stress response
- AARE:
-
Amino acid response element
- ATF4:
-
Activating transcription factor 4
- GCN2:
-
General control nonderepressible 2, or eIF2α kinase 4
- ORF:
-
Open reading frame
References
Dang Do AN, Kimball SR, Cavener DR, Jefferson LS (2009) eIF2α kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol Genomics 38:328–341
Deval C, Chaveroux C, Maurin AC, Cherasse Y, Parry L, Carraro V, Milenkovic D, Ferrara M, Bruhat A, Jousse C, Fafournoux P (2009) Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J 276:707–718
Gabauer F, Hentze M (2004) Molecular mechanisms of translational control. Mol Cell Biol 5:827–834
Guo F, Cavener DR (2007) The GCN2 eIF2α kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab 5:103–114
Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, Wek RC, Cavener DR, McGrath BC, Rudell JB, Koehnle TJ, Gietzen DW (2005) Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science 307:1776–1778
Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6:1099–1108
Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11:619–633
Hinnebusch AG (2000) Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Sonenberg N, Hershey JWB, Mathews MB (eds) Translational control of gene expression. Cold Spring Harbor Laboratory Press, Plainview, NY, pp 185–243
Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acidcontrol of yeast. Annu Rev Microbiol 59:407–450
Ishikawa F, Akimoto T, Yamamoto H, Araki Y, Yoshie T, Mori K, Hayashi H, Nose K, Shibanuma M (2009) Gene expression profiling identifies a role for CHOP during inhibition of the mitochondrial respiratory chain. J Biochem 146:123–132
Jousse C, Bruhat A, Ferrara M, Fafournoux P (2000) Evidence for multiple signaling pathways in the regulation of gene expression by amino acids in human cell lines. J Nutr 130:1555–1560
Kimball SR, Antonetti DA, Brawley RM, Jefferson LS (1991) Mechanism of inhibition of peptide chain initiation by amino acid deprivation in perfused rat liver. Regulation involving inhibition of eukaryotic initiation factor 2 α phosphatase activity. J Biol Chem 266:1969–1976
Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169:425–434
Kubica N, Jefferson LS, Kimball SR (2006) Eukaryotic initiation factor 2B and its role in alterations in mRNA translation that occur under a number of pathophysiological and physiological conditions. Prog Nucleic Acid Res Mol Biol 81:271–296
Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N (2004) The role of autophagy during the early neonatal starvation period. Nature 432:1032–1036
Lee J-I, Dominy JE Jr, Sikalidis AK, Hirschberger LL, Wang W, Stipanuk MH (2008) HepG2/C3A cells respond to cysteine deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol Genomics 33:218–229
Li X, Wang T, Zhao Z, Weinman SA (2002) The ClC-3 chloride channel promotes acidification of lysosomes in CHO-K1 and Huh-7 cells. Am J Physiol Cell Physiol 282:C1483–C1491
Lu SC, Huang HY (1994) Comparison of sulfur amino acid utilization for GSH synthesis between HepG2 cells and cultured rat hepatocytes. Biochem Pharmacol 47:859–869
Lu PD, Harding HP, Ron D (2004a) Translation reinitiation at alternative open reading frames regulate gene expression in an integrated stress response. J Cell Biol 167:27–33
Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, Kaufman RJ, Ron D, Harding HP (2004b) Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J 23:169–179
Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ (2009) CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 182:466–476
Okamoto F, Kajiya H, Toh K, Uchida S, Yoshikawa M, Sasaki S, Kido MA, Tanaka T, Okabe K (2008) Intracellular ClC-3 chloride channels promote bone resorption in vitro through organelle acidification in mouse osteoclasts. Am J Physiol Cell Physiol 294:C693–C701
Padyana AK, Qiu H, Roll-Mecak A, Hinnebusch AG, Burley SK (2005) Structural basis for autoinhibition and mutational activation of eukaryotic initiation factor 2α protein kinase GCN2. J Biol Chem 280:29289–29299
Palii SS, Kays CE, Deval C, Bruhat A, Fafournoux P, Kilberg MS (2009) Specificity of amino acid regulated gene expression: analysis of genes subjected to either complete or single amino acid deprivation. Amino Acids 37:79–88
Pan YX, Chen H, Thiaville MM, Kilberg MS (2007) Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem J 401:299–307
Patwari P, Higgins LJ, Chutkow WA, Yoshioka J, Lee RT (2006) The interaction of thioredoxin with Txnip. Evidence for formation of a mixed disulfide by disulfide exchange. J Biol Chem 281:21884–21891
Sato H, Nomura S, Maebara K, Sato K, Tamba M, Bannai S (2004) Transcriptional control of cystine/glutamate transporter gene by amino acid deprivation. Biochem Biophys Res Commun 325:109–116
Shan J, Ord D, Ord T, Kilberg MS (2009) Elevated ATF4 expression, in the absence of other signals, is sufficient for transcriptional induction via CCAAT enhancer-binding protein-activating transcription factor response elements. J Biol Chem 284:21241–21248
Su N, Thiaville MM, Awad K, Gjymishka A, Brant JO, Yang TP, Kilberg MS (2009) Protein or amino acid deprivation differentially regulates the hepatic forkhead box protein A (FOXA) genes through an activating transcription factor-4-independent pathway. Hepatology 50:282–290
Tallóczy Z, Jiang W, Virgin HW 4th, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B (2002) Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci USA 99:190–195
Thiaville MM, Pan YX, Gjymishka A, Zhong C, Kaufman RJ, Kilberg MS (2008) MEK signaling is required for phosphorylation of eIF2α following amino acid limitation of HepG2 human hepatoma cells. J Biol Chem 283:10848–10857
Vesely PW, Staber PB, Hoefler G, Kenner L (2009) Translational regulation mechanisms of AP-1 proteins. Mutat Res 682:7–12
Wek SA, Zhu S, Wek RC (1995) The histidyl-tRNA synthetase-related sequence in the eIF-2α a protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol 15:4497–4506
Wek RC, Jiang HY, Anthony TG (2006) Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34:7–11
Acknowledgments
We gratefully acknowledge the kind gifts of ATF4 antibody from Dr. Michael S. Kilberg, University of Florida, Gainesville, FL. The microarray analyses were done by the Cornell University microarray core facility center (Cornell Big Red Spots) and were funded in part by the Cornell University Center for Vertebrate Genomics. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases through Public Health Service Grants DK056649 and DK0664303.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sikalidis, A.K., Lee, JI. & Stipanuk, M.H. Gene expression and integrated stress response in HepG2/C3A cells cultured in amino acid deficient medium. Amino Acids 41, 159–171 (2011). https://doi.org/10.1007/s00726-010-0571-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-010-0571-x