Abstract
The activities of a cyclohexene series of influenza neuraminidase inhibitors were studied based on the combination of 3D-QSAR, molecular docking, and molecular dynamics methods. The 3D-QSAR models were established by comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA) methods. The optimum CoMFA and CoMSIA models yielded satisfactory statistical results: the leave-one-out cross-validation correlation coefficients (q 2) were 0.722 and 0.779, respectively. The corresponding non-cross-validated r 2 were both 0.996. Based on the built 3D-QSAR models, several new neuraminidase inhibitor analogs were designed. Molecular docking elucidated the conformations of compounds and key amino acid residues at the docking pocket of neuraminidase protein. Molecular dynamics simulation further determined the binding process and validated the rationality of docking results.
Graphical Abstract

.
Similar content being viewed by others
Introduction
Pandemic influenza outbreaks pose a significant threat to public health as highlighted by the latest emergence of highly pathogenic avian influenza H7N9 [1, 2] and the recent 2009 outbreaks of swine-oriented H1N1 viruses (2009 H1N1) [3]. Currently, prevention and treatment of influenza rely on inactivated vaccines and antiviral drugs. However, attempts to control this disease through immunization have been hampered by the rapidity with which the virus mutates. Therefore, the development of effective and safe antiviral agents is even more important in the event that new highly virulent strains can lead to global pandemics resulting in millions of deaths [4].
Many studies have demonstrated that the influenza virus neuraminidase (NA), a surface glycoprotein located on the virus surface, is a highly successful clinical target for the treatment of influenza infections [5, 6]. Neuraminidase can cleave terminal sialic acid residues from glycoconjugates, which is essential for virus replication and infectivity [7]. It has been postulated that NA is required in the elution of newly synthesized virus from infected cells [8–10]. It may also promote viral movement through respiratory tract mucus, thus enhancing viral infectivity [11]. Therefore, NA has been regarded as an important target for designing agents against influenza viruses.
Based on the NA crystal structures elucidated in the early 1990s, many highly selective NA inhibitors are reasonably designed. A potent inhibitor, zanamivir (Relenza), has been shown to have strong antiviral activity in animal models and in human trials [12, 13]. However, due to poor oral bioavailability, zanamivir is applied topically to the respiratory tract as an intranasal spray or inhalant [12–14]. Aiming at developing oral agents against influenza infection, Kim et al. [15–20] have designed and synthesized a series of carbocyclic NA inhibitors with various lipophilic side chains. GS 4071 ((3R,4R,5S)-4-cetamido-5-amino-3 (1-ethylpropoxy)-1-cyclohexene-1-carboxylic acid) is the most potent in this series with IC 50 of 1 nmol/dm3 [16]. Oseltamivir (GS4104, Tamiflu™), the ethyl ester prodrug of GS4071, a striking influenza virus neuraminidase inhibitor, was developed to enhance oral absorption and increase serum half-life [21]. At present, both zanamivir and oseltamivir are effective inhibitors of both A and B forms of neuraminidase. They have been approved by the FDA for the treatment of influenza. More recently, two other neuraminidase inhibitors, peramivir [22] and laninamivir [23], were also approved as anti-influenza drugs.
Despite their outstanding potency, all these inhibitors have limitations: for example, zanamivir suffers from low oral bioavailability, and oseltamivir is highly vulnerable to inactivation because of viral mutation. In addition, some people using oseltamivir and zanamivir have had rare side effects of sudden confusion, delirium, hallucinations, unusual behavior, or self-injury [24, 25]. Therefore, the drugs currently in use may not fully protect humans, and thus a new generation of antiinfluenza drugs is needed.
To develop new, more effective (more biologically accessible, less toxic, without side effects) NA inhibitors, an effective tool for drug design, the quantitative structure–activity relationship (QSAR) method, had already been applied in this area. For instance, Verma and Hansh [26] developed 17 QSAR models for different sets of compounds including benzoic acid derivatives [27], carbocyclic derivatives [15–20], cyclopentane amide derivative [28], isoquinolines [29], and pyrrolidines [30] to understand chemical–biological interactions governing their activities toward influenza neuraminidase. Two models, topological and geometric, were established to estimate the inhibitory activity of NA inhibitors, which are cyclohexene and cyclopentane derivatives [31]. A 3D-QSAR model was also established by use of descriptors calculated by a holographic vector of the atomic interaction field analysis (HoVAIFA) method [32]. These models can identify some critical structure features for inhibitory activity. However, the traditional 2D method does not take 3D structural features into account and lacks spatial information about compounds [33]. Therefore, comprehensive molecular structure features that contribute to the inhibitory activity of NA inhibitors are still limited. In this work, 3D-QSAR methods, i.e., comparative molecular field analyses (CoMFA) [34] and comparative molecular similarity index analyses (CoMSIA) [35, 36], were applied to gain insights into the key structural factors affecting inhibitory activity of NA inhibitors. The developed models can not only be used to predict the activity of newly designed inhibitors, but also provide beneficial information in structural modifications for designing new inhibitors with desired inhibitory activity. In addition, molecular docking was carried out to study the binding modes of inhibitors at the active site of the NA protein. Molecular dynamics (MD) simulation was performed to confirm the reliability of docking results.
Results and discussion
Data sets
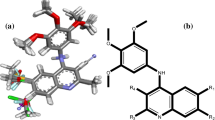
The 35 compounds involved in this study have been reported by the same group [15–20]. The structures and biological activities expressed as pIC 50 against influenza A are shown in Tables 1 and 2. The samples were randomly divided into a training set of 30 compounds for model generation and a test set of five compounds for model validation. The data set compounds were selected by considering both the distribution of biological data and structural diversity. The alignment of training set compounds is shown in Fig. 1.
CoMFA and CoMSIA statistical results
To generate statistically significant 3D-QSAR models, the regression analysis was carried out using the partial least squares (PLS) method [37, 38]. CoMFA and CoMSIA models were developed, and the final models were selected according to the statistical parameters. The statistical results for the final CoMFA and CoMSIA models are summarized in Table 3. PLS analysis on all of the compounds in the training set resulted in a CoMFA model with a cross-validated q 2 of 0.722. This model gave an optimal number of components (ONC) of 10 and a conventional correlation coefficient r 2 of 0.996. The corresponding steric and electrostatic field descriptors explained 72.1 and 27.9 % of the total variance. For CoMSIA analysis, five descriptor fields (steric, electrostatic, hydrophobic, hydrogen bond-donor, and hydrogen bond acceptor) were considered. However, we found that the CoMSIA descriptors such as the steric and hydrogen-bond donor play significant roles in the prediction of inhibitory activity. An excellent value of 0.901 for r 2 prediction was obtained for this model with the q 2 of 0.685. Incorporation of the electrostatic, hydrophobic, or hydrogen bond acceptor field descriptors leads to a small decrease in q 2 (0.609–0.684) and r 2 prediction (0.704-0.855). The CoMSIA model based on hydrophobic and hydrogen bond donor fields was found to have marginally better q 2 of 0.779 with a little drop in the r 2 prediction (0.862). The relationship between actual and predicted pIC 50 value of the training and test set molecules is illustrated in Fig. 2a, b for the CoMFA and CoMSIA models. Herein almost all points are located on the diagonal line.
CoMFA and CoMSIA contour maps
To visualize the field effects on the target compounds in 3D space, the contour maps (Figs. 3, 4, 5) produced by CoMFA (a) and CoMSIA (b) were analyzed by superimposing them onto the most active molecule 22. These contour maps are significant for new drug design, as they show regions in 3D space where modifications of the molecular fields strongly correlated with variations in biological activity.
As shown in Fig. 3, the sterically favorable regions are represented in green and unfavorable regions in yellow. It can be observed that the steric contour map of CoMFA (Fig. 3a) is similar to that of CoMSIA (Fig. 3b). A small green contour covering the ethyl group linked to C1 of R indicates the importance of the presence of a bulky group in this region for biological activity. Thus, compounds 23 and 24 without a bulky group at this position exhibit decreased biological activity. Similarly, the higher pIC 50 values of 9.00 in 18, 21, and 26 are indicative of the importance of a bulky group at this position. The large yellow contour surrounding the aryl ring indicates that compounds with bulky substitution could not possess good biological activity as observed in 24.
The electrostatic field contour maps of CoMFA and CoMSIA are shown in Fig. 4a, b. The electrostatic field is indicated by blue- and red-colored contours, where the blue regions denote that the electropositive groups are favorable to the activity and the red regions indicate that the electronegative groups are favorable to the activity. As shown in Fig. 4a, two pieces of medium-sized region of red contour located at the six-position of cyclohexene ring show the importance of electronegative atoms in imparting better biological activity. This is reflected in the increased biological activity of 28–30 of the training set. Two medium-sized blue contours observed near C3 of R suggest that this position is not suitable for substitution with the electronegative atom. The poor biological activity of 1 is the result of the replacement of H atoms of the methyl group by F atoms. Moreover, it had been recognized that most of cyclohexene ring is encompassed by the red-colored map. This observation demonstrates that these positions are suitable for substitution with electronegative atoms. On the other hand, the appearance can also demonstrate that the carboxyl, amino, and amide groups at the cyclohexene ring are very important for bioactivity.
In hydrophobic fields, yellow and white contours highlight areas where hydrophobic and hydrophilic properties are favored. In Fig. 5a, the yellow contour at C3 of R indicates that this position is suitable for substitution with hydrophobic group. Most of the derivatives involved in this study possess hydrophobic groups at this site, which reveals the importance of the hydrophobic substituent. The white contour at C1 of R indicates that the introduction of hydrophilic moieties at these positions should improve the biological activity. A larger white contour covering the R substituent suggests that the hydrophilic group may be favored. In addition, a small white contour at the ortho position of carboxyl in the cyclohexene ring indicates that the position is suitable for substitution with a hydrophilic group. As shown in Fig. 5b, the purple contours represent the position where the hydrogen bond donor disfavors the biological activity, and the cyan contours show that the presence of donor groups in this region should produce better biological activity. Two pieces of large cyan contours near the region of amide indicate that introduction of hydrogen bond donor moieties should improve the biological activity. This accounts for the better biological activity of 28, 29, and 30. One larger purple contour directed toward the amino group reveals that the hydrogen bond donor substituent at the position is unfavorable to the activity. As shown in Fig. 5c, the hydrogen bond acceptor field is represented by magenta and red contours, in which the magenta contours denote regions where the hydrogen bond acceptor group would be beneficial to the bioactivity, whereas red contours representing the hydrogen bond acceptor group would decrease the bioactivity. Two magenta polyhedrons near the carboxyl of the cyclohexene ring suggest this region is favored for hydrogen bond acceptor interactions. One medium-sized polyhedron located at the C1 of the R substituent group shows disfavored regions for hydrogen bond interactions, suggesting that hydrogen bond acceptor substituent maybe decrease activity.
Molecular design of new NA inhibitors
The detailed contour map analysis of both CoMFA and CoMSIA models empowered us to identify structural requirements for the observed inhibitory activity. Based on QSAR results, inhibitor 22, with the highest activity, was taken as a template to design new compounds. For example, a green contour covering the ethyl group linked to C1 of R indicates the importance of the presence of a bulky group in this region for biological activity. Thus, a bulky isopropyl was introduced to this position, and compound 22a was obtained. A red contour near C6 of the cyclohexene ring shows the importance of the electronegative atom at this position, and thus compound 22b with two F substitutions at C6 was designed. The white contour at the C atom of methylene in ethyl linked to C1 of R indicates that the introduction of hydrophilic moieties at this position perhaps can improve biological activity. So compounds 22c, 22d, and 22e with carboxyl, amino, and hydroxyl substitutions were designed. The structures of compounds 22a-22e are shown in Table 4. Their computed total energies, zero-point energies (ZPE), relative energies (with ZPE corrections), and number of imaginary frequencies are also listed in Table 4. Harmonic vibrational frequency calculations indicate that all isomers 22a-22e are local minima on their potential energy surfaces at the B3LYP level of theory. To predict their biological activity, the CoMFA and CoMSIA models were applied to these new molecules; the corresponding results are listed in Table 5. The results show that the pIC 50 values of these compounds are all higher than 6.918, indicative of their good biological activity. Based on the CoMFA model, 22a, 22d, and 22e have higher pIC 50 values than that of the most active molecule 22, so we predict these three compounds perhaps should be regarded as good candidates for experimental synthesis.
Docking analysis
A number of high-resolution crystal structures of influenza NA and its complex with various small molecule inhibitors have been determined and are available from the Protein Databank. Based on the analysis of these structures, Kim et al. [18] revealed that electrostatic interactions might play a critical role for any successful inhibitors. Herein two new most potent analogs, 22a and 22d, are selected for more detailed analysis. Figure 6 shows the interacting mode of compounds 22a and 22d in the binding site of the NA receptor. Some key residues, such as Arg292, Arg371, Arg152, Tyr406, and Trp178, as well as hydrogen bonds between the selected compound and the residues were also labeled. As shown in Fig. 6a, compound 22a was docked in the binding cavity with the carboxyl directing toward the hydrophilic group of Arg292, Arg371, and Tyr406. The ligand is anchored in the binding site perhaps via five H-bonds. The carboxyl oxygen atom of 22a perhaps acts as an acceptor to form two hydrogen bonds with the H atom of the –NH2 groups of the Arg292 residue and one hydrogen bond with the H atom of the –NH2 group of the Arg371 residue. It may also form one hydrogen bond with the H atom of the –OH group of Tyr406. Another hydrogen bond is formed between the amide oxygen atom of the cyclohexene ring and hydrogen atom of the –NH2 group in Arg152 residue. Indeed, the crucial electrostatic interactions between the NA protein and residues Arg118 and Glu119 were observed in the binding pocket. Figure 6b depicts the docking result of compound 22d. This ligand was docked in the binding cavity with the amide directing toward the hydrophilic group of Arg292, Arg371, and Tyr406. The ligand is anchored in the binding site perhaps via six H-bonds. The amide oxygen atom perhaps acts as an acceptor to form three hydrogen bonds with H atoms of the –NH2 group in Arg292 and Arg371 residues and one hydrogen bond with H atom of the –OH group in Tyr406 residue. Another hydrogen bond is formed between the carboxyl oxygen atom of Glu119 and H atom of the –NH2 group in the cyclohexene ring. The sixth hydrogen bond is formed between the carboxyl oxygen atom of Trp178 and H atom of the –NH2 group in the R substitute. Similarly, the crucial electrostatic interactions between the NA protein and residues Arg152, Glu227, and Arg118 were also found in the binding pocket.
MD simulations of complexes
The MD simulations of the two above-mentioned docking complexes (22a-NA and 22d-NA) were carried out for 1,200 ps to validate the dynamic stability of the two systems. The superposition of the average structure of the last 200-ps MD simulation and the initial docked structure is shown in Fig. 7. Where the magenta ligand and ribbon represent the average structure and the corresponding MD complex, and the green ligand and ribbon represent the initial structure and the corresponding docked complex. As shown in both Fig. 7a, b, it can be recognized that the average structure extracted from MD simulations, and the initial docked structure of the complex is in the same binding pocket. Except for a slight drift and rotation of bonds, there seems to be no significant difference between the average and the initial docked structure of the complex. It can be inferred that the binding pocket and the conformation of the ligand are stable, and the docking results are reliable.
Conclusion
CoMFA and CoMSIA studies on 35 cyclohexene-based NA inhibitors were carried out to develop 3D-QSAR models that provided good internal and external predictivity. The resulted models can be extrapolated to predict novel and more potent molecules. The contour maps obtained from the CoMFA and CoMSIA analysis could guide the design of new chemical entities with high NA inhibitory activity. Based on the built QSAR models, several novel NA inhibitors were designed, and the best candidates for experimental synthesis were suggested. To study the binding modes of inhibitors at the active site of NA protein, molecular docking studies of representative compounds were performed. Some key residues such as Arg292, Arg371, Arg152, Tyr406, and Trp178 as well as hydrogen bonds between the selected compound and the residues were found. To further confirm the reliability of docking results, MD simulations were carried out for representative compounds. We hope our research may provide a basis for the development of new NA inhibitors.
Materials and methods
Molecular modeling and database alignment
The molecular modeling and 3D-QSAR studies were performed using the molecular modeling package SYBYL-X 2.0 (Tripos, Inc., USA). Three-dimensional structures of all compounds were constructed by using the Sketch Molecule module. Energy minimization was performed by the Powell gradient algorithm with the Tripos force field [39] and Gasteiger-Hückel charge [40]. The maximum iterations for the minimization were set to 10,000. The minimization was terminated when the energy gradient convergence criterion of 0.021 kJ/mol Å was reached [32].
One of the critical steps in 3D-QSAR studies is the selection of active conformation and alignment of molecules. The success of these methods strongly depended on the relative position of the ligands in the fixed lattice before the generation of 3D descriptors. The database alignment method was performed. The fragment used for the alignment is the cyclohexene carbon ring, and the most active molecule 22 is used as a template.
CoMFA and CoMSIA modeling
CoMFA steric and electrostatic interaction fields were calculated at each lattice intersection on a regularly spaced grid of 3 Å. The grid pattern was generated automatically by the SYBYL/CoMFA routine. As a probe atom, an sp3-hybridized carbon atom of +1.0 charges with a van der Waals radius of 1.52 Å was used at each intersection [41]. For the CoMFA analysis, two descriptors, steric and electrostatic fields were generated and scaled by the CoMFA-STD method with default energy of 125.4 kJ/mol. Steric interactions were calculated using the Lennard-Jones 6–12 potential, while electrostatic interactions were calculated using the Coulomb potential. In the case of CoMSIA analysis, similarity index descriptors were derived with the same lattice box that was used in CoMFA. Five fields, steric, electrostatic, hydrophobic, hydrogen bond donor, and hydrogen bond acceptor interactions, were calculated using the same probe atom as for the CoMFA analysis.
Regression analysis and model validation
The CoMFA and CoMSIA descriptors were used as independent variables, and pIC 50 values were used as the dependent variables. The performance of models was evaluated using the leave-one-out (LOO) cross-validation method. The optimal number of components (ONC) equal to that yielding the highest cross-validated q 2 was used to generate the final PLS regression models. The conventional correlation coefficient r 2, standard error of estimate (SEE), and F ratio between the variances of experimental and predicted activity values were then computed for the final PLS models. The CoMFA and CoMSIA results were interpreted graphically by the contribution maps using the field type “PLSstdev × PLScoeff”. To validate the CoMFA- and CoMSIA-derived models, the predictive ability \( r_{\text{pred}}^{2} \) was determined for the test set molecules. The cross-validated correlation coefficient q 2 and \( r_{\text{pred}}^{2} \) were calculated by using Eqs. (1) and (2),
where Y exp, Y pred, and Y mean are the experimental, predicted, and mean values of activity.
Herein PRESS means the sum of squared deviations between experimental and predicted activity values for each molecule in the test set. SD means the sum of squared deviations between the experimental activities of the compounds in the test set and the mean activity of the training molecules [33].
Computational methods
The calculations for the designed new molecules were performed using the Gaussian 03 program package [42]. We optimized geometries and calculated the harmonic vibrational frequencies at the B3LYP/6-31G* level of theory, where B3LYP is the DFT method using Becke’s three-parameter gradient-corrected functional [43] with the gradient corrected correlation of Lee et al. [44], and 6–31G* is the used basis set [45]. Stationary points were characterized as minima without any imaginary vibrational frequency.
Molecular docking
To study the binding mode of the inhibitors in the active site of NA protein, molecular docking was performed using the Surflex-Dock module in SYBYL-X 2.0. The crystal structure of the NA receptor complex was retrieved from the RCSB Protein Data Bank (PDB entry code: 4K1K) [46]. The ligands were docked in the corresponding protein’s binding site by an empirical scoring function and a patented search engine in Surflex-Dock. Before the docking process, one natural ligand was extracted; the other natural ligands and water molecules were removed from the crystal structure. Subsequently, the protein was prepared by using the Biopolymer module implemented in Sybyl. The polar hydrogen atoms were added, and Gasteiger-Hückel charges were assigned to protein atoms. The automated docking manner was applied in the present work. Other parameters were established by default in the software. Surflex-Dock total scores, which were expressed in −log10(K d) units to represent binding affinities, were applied to estimate the ligand-receptor interactions of newly designed molecules.
Molecular dynamics (MD) simulations
To confirm the docking results, the MD simulations [47–49] were carried out in SYBYL-X 2.0 software. The docked complexes of NA protein with two designed most active molecules are used as initial conformations. The system setup for simulation included an 8-Å cutoff for non-bonded van der Waals interactions and periodic boundary conditions. Constant temperature (300 K) and volume were maintained with the time constant for a heat bath coupling of 100 fs. The time step of 1 fs was used to integrate the equations of motion, and the snapshot time was 100 fs. The Boltzmann initial velocity was used to start the simulation. Other parameters were set by default in Sybyl.
At the beginning of the production-run phase, the whole system was first subjected to a gradual temperature change from 319 to 294 K. The whole system was equilibrated for 600 ps, followed by another 600 ps of the molecular dynamics production phase. The resulting trajectories were analyzed by the Analyze module of Sybyl.
References
Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xing N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y (2013) N Engl J Med 368:1888
Uyeki TM, Cox NJ (2013) N Engl J Med 368:1862
Liao HY, Hsu CH, Wang SC, Liang CH, Yen HY, Su CY, Chen CH, Jan JT, Ren CT, Chen CH, Cheng TR, Wu CY, Wong CH (2010) J Am Chem Soc 132:14849
Cui Y, Jiao Z, Gong J, Yu Q, Zheng X, Quan J, Luo M, Yang Z (2010) Org Lett 12:4
Schmidt AC (2004) Drugs 64:2031
De Clercq E (2006) Nat Rev Drug Discov 5:1015
Mendel DB, Tai CY, Escarpe PA, Li W, Sidwell RW, Huffman JH, Sweet C, Jakeman KJ, Merson J, Lacy SA, Lew W, Williams MA, Zhang L, Chen MS, Bischofberger N, Kim CU (1998) Antimicrob Agents Chemother 42:640
Liu C, Eichelberger MC, Compans RW, Air GM (1995) J Virol 69:1099
Palese P, Compans RW (1976) J Gen Virol 33:159
Palese P, Tobita K, Ueda M, Compans RW (1974) Virology 61:397
Klenk HD, Rott R (1988) Adv Virus Res 34:247
Ryan DM, Ticehurst J, Dempsey MH (1995) Antimicrob Agents Chemother 39:2583
Ryan DM, Ticehurst J, Dempsey MH, Penn CR (1994) Antimicrob Agents Chemother 38:2270
Hayden FG, Treanor JJ, Betts AF, Lobo M, Esinhart JD, Hussey EK (1996) JAMA 275:295
Kim CU, Lew W, Williams MA, Wu H, Zhang L, Chen X, Escarpe PA, Mendel DB, Laver WG, Stevens RC (1998) J Med Chem 41:2451
Kim CU, Lew W, Willams MA, Liu H, Zhang L, Swaminathan S, Bischofberer N, Chen MS, Mendel DB, Tai CY, Laver WG, Stevens RC (1997) J Am Chem Soc 119:681
Lew W, Escarpe PA, Mendel DB, Sweeny D, Kim CU (1999) Bioorg Med Chem Lett 9:2811
Williams MA, Lew W, Mendel DB, Tai CY, Escarpe PA, Laver WG (1997) Bioorg Med Chem Lett 7:1837
Lew W, Wu H, Mendel DB, Escarpe PA, Chen X, Laver WG, Graves BJ, Kim CU (1998) Bioorg Med Chem Lett 8:3321
Lew W, Wu H, Chen X, Graves BJ, Escarpe PA, MacArthur HL, Mendel DB, Kim CU (2000) Bioorg Med Chem Lett 10:1257
Burger RA, Billingsley JL, Huffman JH, Bailey KW, Kim CU, Sidwell RW (2000) Immunopharmacology 47:45
Jain S, Fry AM (2011) Clin Infect Dis 52:707
Kubo S, Tomozawa T, Kakuta M, Tokumitsu A, Yamashita M (2010) Antimicrob Agents Chemother 54:1256
Hata K, Koseki K, Yamaguchi K, Moriya S, Suzuki Y, Yingsakmongkon S, Hirai G, Sodeoka M, Von Itzstein M, Miyagi T (2008) Antimicrob Agents Chemother 52:3484
Kitching A, Roche A, Balasegaram S, Heathcock R, Maguire H (2009) Eurosurveillance 14:2
Verma RP, Hansch C (2006) Bioorg Med Chem 14:982
Chand P, Babu YS, Bantia S, Chu N, Cole LB, Kotian PL, Laver WG, Montgomery JA, Pathak VP, Petty SL, Shrout DP, Walsh DA, Walsh GM (1997) J Med Chem 40:4030
Chand P, Babu YS, Bantia S, Rowland S, Dehghani A, Kotian PL, Hutchison TL, Ali S, Brouillette W, EI-Kattan Y, Lin TH (2004) J Med Chem 47:1919
Tute MS (1970) J Med Chem 13:48
Wang GT, Chen Y, Wang S, Gentles R, Sowin T, Kati W, Muchmore S, Giranda V, Stewart K, Sham H, Kempf D, Laver WG (2001) J Med Chem 44:1192
Tsygankova IG, Zhenodarova SM (2006) Russ J Gen Chem 76:1618
Sun J, Cai S, Yan N, Mei H (2010) Eur J Med Chem 45:1008
Li X, Ye L, Wang X, Wang X, Liu H, Zhu Y, Yu H (2012) Toxicol Appl Pharm 265:300
Cramer RD III, Patterson DE, Bunce JD (1988) J Am Chem Soc 110:5959
Klebe G, Abraham U, Mietzner T (1994) J Med Chem 37:4130
Böhm M, Stürzebecher J, Klebe G (1999) J Med Chem 42:458
Ståhle L, Wold S (1987) J Chemom 1:185
Wold S, Albano C, Dunn WJ, Edlund U, Esbenson K, Geladi P, Hellberg S, Lindburg W, Sjostrom M (1984) Mathematics and statistics in chemistry. Reidel, Dordrecht
Clark M, Cramer RD III, Opdenbosch NV (1989) J Comput Chem 10:982
Gasteiger J, Marsili M (1980) Tetrahedron 36:3219
Zhang QY, Wan J, Xu X, Yang GF, Ren YL, Liu JJ, Wang H, Guo Y (2007) J Comb Chem 9:131
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, AlLaham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2003) Gaussian 03, Revision B.04. Gaussian Inc, Pittsburgh
Becke AD (1993) J Chem Phys 98:1372
Lee C, Yang W, Parr RG (1988) Phys Rev B 37:785
Hehre WJ, Radom L, Schleyer PVR, Pople JA (1986) Ab initio molecular orbital theory. Wiley, New York
Wu Y, Qin G, Gao F, Liu Y, Vavricka CJ, Qi J, Jiang H, Yu K, Gao GF (2013) Sci Rep 3:1
Kai ZP, Ling Y, Liu WJ, Zhao F, Yang XL (2006) Biochim Biophys Acta 1764:70
UI-HAq Z, Khan W, Zia SR, Iqbal S (2012) J Mol Graph Model 36:48
Yuan M, Luo M, Song Y, Xu Q, Wang X, Cao Y, Bu X, Ren Y, Hu X (2011) Bioorg Med Chem 19:1189
Acknowledgments
This research was supported by the Natural Science Foundation of China (No. 21172148), Innovation Program of Shanghai Municipal Education Commission (No. 11YZ224), and Curriculum Building Program of Graduate Student Department of Shanghai Institute of Technology (No. 101YM120003) and Shanghai Leading Academic Discipline Project, Project No. J51503.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cheng, L.P., Huang, X.Y., Wang, Z. et al. Combined 3D-QSAR, molecular docking, and molecular dynamics study on potent cyclohexene-based influenza neuraminidase inhibitors. Monatsh Chem 145, 1213–1225 (2014). https://doi.org/10.1007/s00706-014-1176-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00706-014-1176-y