Abstract
Nyamanini virus (NYMV) and Midway virus (MIDWV) are unclassified tick-borne agents that infect land birds and seabirds, respectively. The recent molecular characterization of both viruses confirmed their already known close serological relationship and revealed them to be nonsegmented, single- and negative-stranded RNA viruses that are clearly related to, but quite distinct from, members of the order Mononegavirales (bornaviruses, filoviruses, paramyxoviruses, and rhabdoviruses). A third agent, soybean cyst nematode virus 1 (SbCNV-1, previously named soybean cyst nematode nyavirus), was recently found to be an additional member of this new virus group. Here, we review the current knowledge about all three viruses and propose classifying them as members of a new mononegaviral family, Nyamiviridae.
Similar content being viewed by others
Introduction
The viral order Mononegavirales was created in 1991 to include the already well-established families Paramyxoviridae, Rhabdoviridae, and Filoviridae [46] and was expanded in 1997 by inclusion of the family Bornaviridae [47]. All mononegaviruses are characterized by (1) having a linear, monopartite, single-stranded RNA genome of negative polarity, (2) having a similar genomic organization in the order 3′-untranslated region (UTR) – core protein genes – envelope protein genes – RNA-dependent RNA polymerase gene – 5′-UTR, (3) transcription of discrete mRNAs by sequential interrupted synthesis from a single promoter, (4) synthesis of a complete RNA antigenome during replication, and (5) the formation of virions whose envelopes are derived from the host cell. The members of the order are assigned to the four established families based on genome size, coding capacity, virion morphology, host range, and pathogenicity as well as by phylogenetic comparison of the core regions of their polymerases (domain III) [19, 46, 47]. The order has grown considerably since its establishment, and currently includes more than 18 genera and 98 species. In addition, numerous viruses have been identified as definite members of the order, and often as members of particular families, but have not yet been assigned to genera and/or species because of a lack of sequence information or insufficient biological characterization [2, 19, 32].
Recent breakthroughs in sequencing technologies indicate that the virus sphere is considerably larger than that currently acknowledged by the International Committee on Taxonomy of Viruses (ICTV) classification framework. The latest, 9th ICTV Report lists 2,284 virus and viroid species, 349 genera, 19 subfamilies, 87 families, and 6 orders [32]. At the same time, investigators have reached a common consensus that the vast majority of the millions of distinct living organisms likely carry one or more distinct viruses [6, 64]. Several studies clearly support this view. For instance, some 20 viral metagenomic surveys, i.e., shotgun sequencing of purified virion populations collected from different environments, have demonstrated that dominant virus sequences detected in these samples are rarely represented by cultured viruses acknowledged by the ICTV or in current databases [50]. More than half of the detected viral sequences during such studies are completely novel. The remainder of the viral sequences are mostly only distantly related to known sequences and therefore most likely represent viruses requiring assignment to novel taxa [17, 49, 66]. Importantly, improving bioinformatics capabilities have repeatedly allowed assembly of the complete genomes of multiple previously unknown and possibly nonculturable viruses [15, 18, 22, 45, 48, 53, 60].
Accurate taxonomic classification of viruses that have been cultured and sequenced but not further characterized is inherently needed. More importantly, we believe viruses should be classified based on detection and sequencing of nearly complete viral genomic nucleic acids from defined biological environments (niches). First, due to the vast number of viruses in nature, many viruses will probably not be characterized in detail in the laboratory, especially when their importance for public, animal, and crop health or their economic impact is unknown a priori. Second, reminiscent of problems in bacteriology, it is highly unlikely that the majority of viruses can ever be cultured, as they may depend on particular host cells, nutrients, or environmental factors that cannot be mimicked in the laboratory. At the same time, novel, seemingly unimportant viruses may express proteins of appreciable scientific, medical, or economic interest, hold clues with regard to viral evolution and ecology, or may be identified as pathogens by indirect means. Consequently, proper cataloguing of these novel viruses in databases and their proper phylogenetic classification in taxonomy frameworks is necessary [36, 50]. ICTV’s current International Code of Virus Classification and Nomenclature (ICVCN) does not prohibit the classification of viruses for which isolates are not available, but clearly, a minimal set of biological information about a nonculturable virus should be available for classification. Unfortunately, such minimal datasets have yet to be established for viral classification. On the other hand, the ICTV has classified hundreds of viruses for which there are isolates but which are basically uncharacterized (e.g., by sequencing) except for serological cross-reaction tests. We think that in the absence of other data, the information about a virus gleaned from an available full-length genome can be as informative, or maybe more informative, for classification purposes than a serological cross-reaction test or the availability of an isolate.
Results from recent studies have evoked a need to discuss and establish novel classification criteria for uncharacterized and/or uncultured mononegaviruses. For instance, the full genome sequence of a clearly novel filovirus, Lloviu virus, was determined in the absence of virus culture in tissues of deceased fruit bats in Spain [45]. A novel, clearly distinct rhabdovirus, Bas-Congo virus, was identified by determination of its full-length genome in sera obtained from humans with acute-phase viral hemorrhagic fever [22]. Most impressively, results from an extensive sequencing study in bats and rodents performed by Drexler et al. has recently revealed at least 66 new members of the family Paramyxoviridae in the absence of virus isolates [18]. In further work, nearly complete genome sequences of several of these viruses were determined, and phylogenetic analysis using detected genome fragments suggested that each of the 66 new viruses could represent a new paramyxovirus species [18]. Together, these data indicate that mononegavirus diversity is underappreciated, that many mononegaviruses may only be discovered by sequencing efforts, and that the current mononegavirus classification criteria may be inadequate. To avoid appreciable classification backlogs, the mononegavirus ICTV Study Groups may need to become more proactive and more flexible regarding mononegavirus classification.
Here, we address the classification of three mononegaviruses that, according to the currently valid mononegavirus classification criteria [19, 32, 46, 47], cannot be assigned to any of the four established mononegavirus families. We therefore propose, based on sequence and biological data, to classify these viruses as members of a new mononegaviral family, Nyamiviridae.
Nyamanini virus
On November 19, 1957, B. M. McIntosh collected samples that contained a novel virus from a cattle egret (Animalia: Aves: Pelicaniformes: Ardeidae: Bulbulcus ibis ibis Linnaeus, 1758) at Nyamanini Pan in the Ndumu Game Reserve, northern Natal, Union of South Africa (today the Republic of South Africa). McIntosh re-isolated the virus four times between 1959 and 1960 from cattle egrets at Naboomspruit and 14 times from associated soft ticks (Animalia: Arthropoda: Arachnida: Ixodida: Argasidae: Argas (Persicargas) arboreus Kaiser, Hoogstraal & Kohls, 1964) and named it Nyamanini virus. McIntosh then proceeded to register this virus in the “Catalogue of Arthropod-borne Viruses” in February of 1960 [1, 24, 27, 28, 30, 57] (prototype isolate SAAn2526). Nyamanini virus was also isolated
-
from A. arboreus ticks collected in an egret rookery (isolate Ar1156 collected on January 29, 1954 and Ar1304 collected on August 12, 1954) in the Nile Delta of the Republic of Egypt (today the Arab Republic of Egypt) and in a sample from a nestling cattle egret (isolate An2252 collected on June 14, 1954) [56, 57];
-
from A. arboreus ticks collected between March 1963 and February 1964 at a heron rookery at Delta Barrage, Qalyūb, Qalyūbīya Governorate, Arab Republic of Egypt (isolate 2558-59 of June 1963 and 75 additional, unnamed, isolates). In 1963, the percentage of infected ticks was found to be highest in June and lowest in September and December [29];
-
from ticks (Argas (Persicargas) robertsi Hoogstraal, Kaiser & Kohls, 1968) collected in 1967 from under the bark of trees in an Asian openbill (Aves: Ciconiiformes: Ciconiidae: Anastomus oscitans Boddaert, 1783) rookery at Wat Phai Lom, Pathum Thani, Thailand [24, 25] (isolate tick39);
-
from A. arboreus ticks (males, females, and nymphs, but not larvae) collected on October 14-15, 1970, in Dalori (11 isolates) and on October 17, 1970, in Gangaba (4 isolates), North Eastern State, Nigeria, from under the bark of trees frequented by cattle egrets [31, 63];
-
from A. robertsi ticks collected in 1972 from Mahadan and Kumbuk trees with nests of eastern grey herons (Aves: Pelicaniformes: Ardeidae: Ardea cinerea rectirostris Gould, 1843), Asian openbills, and cormorants (Phalacrocorax Brisson, 1760 spp.) in Wilpattu Nature Reserve (today Wilpattu National Park), Kumbukwila, Anuradhapura District, North Central Province, Ceylon (now Sri Lanka) [24, 25] (isolate HH21363);
-
from a grey heron in India at an unknown date at an unknown location [25, 59] (isolate I64434); and
-
from ticks (Argas (Aragas) reflexus Fabricius, 1794) collected in Nepal at an unknown date at an unknown location [59] (isolate NepAr89).
Nyamanini virus is not known to cause disease in humans or wild animals. Initial studies failed to detect complement-fixation antibodies to isolate EgAr1304 in 120 acute and 48 convalescent sera of children with fever collected between June and October of 1968 at a hospital in Alexandria, Egypt [43]. Follow-up studies also failed to detect antibodies in 191 human sera collected in Lower Egyptian governorates [16]. Low titers of antibodies against the virus could be detected in 3 of 403 (0.01 %) humans, an unspecified number of goats, 0 of 76 cattle and sheep, and 1 of 9 (0.11 %) donkeys in South Africa [30, 31] and in 3 of 108–109 (2.80 %) buffalo in a Cairo abattoir, 1 of 137 (0.73 %) camels in a Cairo abattoir, and 1 of 101 (0.99 %) stray dogs sampled in the Greater Cairo area. Antibodies could not be detected in 100 sheep, 101 pigs, 187–197 donkeys (numbers vary depending on source), or 94 unspecified rodents [16, 30]. In the laboratory, Nyamanini virus isolate SAAn2526 killed suckling mice within 7–8 days after intracerebral (i.c.) inoculation; some weaned mice also succumbed to infection [30]. Adult guinea pigs and rabbits had an antibody response, but no illness, after i.c. inoculation. Viremia was detected in adult “vervet monkeys” (Chlorocebus aethiops?) injected subcutaneously (s.c.), whereas 6-month-old lambs and 1-day-old chickens only had antibody responses after s.c. and intramuscular (i.m.) injections, respectively. Intravenous injection into a nestling cattle egret and an adult unspecified coot resulted in viremia. Yolk sac injection into an embryonated 8-day-old chicken egg resulted in death 4 days after injection [30]. Isolate 2558-59 killed suckling mice within 7-8 days after i.c. inoculation. Three-week-old mice or half-day-old chicks were not susceptible to Nyamanini virus infection via i.c. or intraperitoneal (i.p.) infection, nor were 5-week-old chickens susceptible after s.c. injection [29]. Likewise, 5-week-old C57/BL6 laboratory mice did not develop symptoms after infection with Nyamanini virus isolate tick39 [23].
Midway virus
On July 12 and August 6, 1966, viruses serologically related to, but distinct from, Nyamanini virus were isolated from ticks (Ornithodoros (Alectorobius) capensis Neuman, 1901) collected from nests of a sooty tern colony (Aves: Charadriiformes: Sternidae: Onychoprion fuscatus Linnaeus, 1766) on Midway Atoll and from nests of unspecified larid birds on Kure Atoll, Hawaii, USA, respectively [13, 14, 55, 67, 68] (isolates RML47153 and RML47156). Other isolations were made
-
between June 22 and 23, 1971, from O. capensis ticks collected from nests of black-tailed gulls (Aves: Charadriiformes: Laridae: Larus crassirostris Vieillot, 1818) on Aomatsushima Islet, Iwate Prefecture, Japan [54, 55] (isolate Cap15);
-
on June 6 and June 20, 1972, from the same samples [54, 55] (isolates Cap39, Cap40, Cap41, Cap43, and Cap44); and
-
on November 10, 1973, from ticks (Ornithodoros (Alectorobius) denmarki Kohls, Sonenshine & Clifford, 1965) collected on Manana Islet, Hawaii, USA [14, 55, 67, 68] (isolate RML 63668).
All of these isolates proved to be serologically identical and therefore are now considered isolates of a distinct virus, Midway virus [14, 54, 55, 67, 68]. Midway virus is not known to cause disease in humans or wild animals, but neutralizing antibodies have been detected in nestlings of black-tailed gulls and black-crowned night herons (Aves: Pelicaniformes: Ardeidae: Nycticorax nycticorax Linnaeus, 1758). However, the fourth suckling Swiss mouse brain passage of isolate Cap15 killed suckling Swiss mice, but not 4-week-old Swiss mice, after i.c., but not after i.p., injection 6–15 days after inoculation. Isolate RML47153 behaved similarly, but also caused some deaths when inoculated i.p. into adult guinea pigs. Hamsters (9 days old) and wet chicks proved resistant to isolate RML47153 inoculation irrespective of the route of inoculation [55].
Soybean cyst nematode nyavirus
In 2011, Bekal et al. published the genomic sequence of a novel Midway virus-related agent detected on January 28, 2010, in a highly inbred laboratory culture of a plant-parasitic nematode (strain Terry Niblack (TN)10), the soybean cyst nematode (Animalia: Nematoda: Chromadorea: Tylenchida: Heteroderidae: Heterodera glycines Ichinohe, 1952). Importantly, the new virus was detected in the surface-sterilized developmental stage of the nematode that hatches from the egg and can move in the soil seeking a soybean plant to parasitize (second-stage juveniles, J2), thereby largely excluding fungal virus contamination [3].
Current classification and nomenclature
Nyamanini virus was not listed in the 1st through 5th Reports of the ICTV [20, 21, 37, 38, 65] despite the publication of its discovery and naming in 1960 [57]. Midway virus was mentioned in the literature for the first time in 1968 as an unnamed virus [13], but the first thorough description was published in 1982 [55]. Not surprisingly, Midway virus was not mentioned in the 4th ICTV Report (1982) [38], but it was also absent from the 5th-8th ICTV Reports (1991-2005) [8, 21, 40, 44]. In the 6th, 7th, and 8th ICTV Reports (2005), Nyamanini virus was mentioned as a virus not yet assigned to any taxon [8, 39, 40]. The most current, 9th ICTV Report (2011), lists both Nyamanini virus and Midway virus as unassigned, but related, viruses that most likely “form a distinct lineage in the order Mononegavirales” [1]. The discovery of the soybean cyst nematode virus [3] (2012) followed the publication of the 9th ICTV Report. Consequently, the virus cannot be found in any of the standard virus taxonomical reference works.
The confirmation of the antigenic relationship between Nyamanini virus and Midway virus by cross-box complement fixation tests resulted in the establishment of the Nyamanini serocomplex, which was abbreviated “NYM” by several authors and “nym” by Cerny [7, 11, 12, 14, 24, 42]. “NYM” was first used as the abbreviation for Nyamanini virus by Hoogstraal in 1974 [25], by McIntosh in 1975 [5], and by Jupp and McIntosh in 1981 [27]. In recent publications, including the latest ICTV Reports, “NYMV” and “MIDWV” have been used as the abbreviations for Nyamanini virus and Midway virus, respectively [1, 3, 8, 23, 42]. Bekal et al. named the MIDWV-related agent they found in soybean cyst nematodes “soybean cyst nematode nyavirus” and abbreviated it ScNV [3]. At the time of this writing, GenBank lists the same virus under the name “soybean cyst nematode midway virus” [GenBank accession #HM849038]. We propose here to rename this virus “soybean cyst nematode virus 1 (SbCNV-1)” so that possible future taxon reorganizations do not require a virus name change, and because the abbreviation “ScNV” is already in use for Saccharomyces 20S RNA narnavirus [33].
Virological and molecular characterization
Cell culture
NYMV (isolate 2558-59) replicates and causes visible cytopathic effects (CPE) by day 5 after inoculation in duck embryo tissue cultures. Plaques are “irregular with incomplete necrosis within their areas and with diffused necrosis on the periphery, giving an opaque appearance” [29]. Isolate tick39 can be propagated in Vero cells [23, 42]. Isolate SAAn2526 causes plaques in LLC-MK2 cells, and isolate EgAr1304 replicates in BHK-21 cells with visible CPE [30].
MIDWV (isolate RML47153, mouse passage 4) replicates in RML-14 tick cells (derived from Dermacentor parumapertus Neumann, 1901) [7], is cytopathic for BHK-21 cells, and produces plaques in Vero cells [42, 55]. The Japanese MIDWV (Hirota) isolates can also be grown in BHK-21 cells, as well as in LLC-MK2 cells [54]. At this time, there is no isolate of SbCNV-1 [3].
Virions
NYMV and MIDWV particles were at first reported to be unusually large, with average diameters of 220-450 nm [9, 55]. More recent studies, utilizing thin-section electron microscopy, suggest the spherical particles to be closer to ≈100-130 nm in diameter [42]. Both NYMV (isolate SAAn2526) and MIDWV (RML47153 and Hirota isolate) particles proved to be sensitive to lipid solvents, such as diethyl ether and sodium deoxycholate, indicating that these particles are enveloped [55]. Electron microscopic studies indicate that the envelope is acquired when the particles bud from the host cell plasma membrane [42]. NYMV (isolate 2558-59) and MIDWV (isolate Cap15) do not hemagglutinate goose erythrocytes (tested pH range, 5.7-6.8 and 6-7, respectively) [29, 55]. MIDWV (isolate RML47153) particles are inactivated within 30 min at 56°C [55], and both NYMV (isolate EgAr1304) and MIDWV (isolate RML47153) particles are inactivated rapidly at pH = 3 [55].
Genomes
NYMV (isolate EgAr1304) and MIDWV (RML47153 and Hirota isolates) replicate in the presence of DNA inhibitors (e.g., 5-bromodeoxyuridine), indicating that their genome consists of RNA and that replication is not dependent on DNA intermediates [55]. The NYMV (isolate tick39) genome is mono-phosphorylated at its 5′ end and contains two nucleotides at its extreme 3′ terminus that are not complementary to the extreme 5′ terminus. The lack of complementary nucleotides prevents perfect panhandle formation of the viral genome (protruding 3′ terminus). These special features of the viral genome appear to facilitate viral evasion of the host-cell interferon response [23]. Genomic sequences are now available for NYMV (isolate tick39), MIDWV (isolate RML47153), and SbCNV-1 (GenBank accession nos. FJ554526, FJ554525, and HM849038, respectively) [3, 42].
The complete NYMV and nearly complete MIDWV genomes are 11,631 nt and ≈11,752 nt long, respectively. The genomes of both viruses contain six obvious open reading frames (ORFs) (Table 1) [42]. The NYMV and MIDWV transcription initiation motifs are 3′-AGUUGG(G/A)(G/A)GAA-5′ and 3′-GGUUGG(G/A)GGG(G/A)-5′, and the transcription termination motifs are 3′-AGAAAUGUUUUU-5′ and 3′-AGAA(U/A)UGUUUUU-5′, respectively [42]. When compared to each other, the lowest similarity score of NYMV vs. MIDWV ORF expression products is 54 %, emphasizing the close relationship of these two viruses (Table 1).
Sequence similarity and structural properties of NYMV and MIDWV suggest that ORF1, 5, and 6 encode the nucleoprotein (N), the glycoprotein (G), and the viral polymerase (L), respectively. The amino acid sequence of the ORF1 expression product has detectable similarity to the nucleoprotein (N) of Borna disease virus (BDV; family Bornaviridae, genus Bornavirus, species Borna disease virus). The amino acid sequence of the ORF6 expression product is similar in sequence to mononegavirus L proteins, suggesting that ORF6 encodes the RNA-dependent RNA polymerase (RdRp). ORF5 most likely encodes the glycoprotein, as it is similar to other mononegaviral G/GP genes in that it encodes an obvious signal peptide and a protein with the typical organization of a type I transmembrane protein, together with multiple glycosylation sites [42].
The NYMV ORF2-derived protein enhances virion-like particle (VLP) formation when co-expressed with ORF4 and ORF5, suggesting that ORFs 2 and 4 encode two distinct matrix proteins, reminiscent of filoviral VP24/VP40 [23]. More recent experiments revealed that a mutant NYMV lacking ORF2 cannot be recovered using reverse genetics, whereas a mutant NYMV lacking ORF4 remains replication-competent. Interestingly, ORF4-deficient NYMV readily established a persisting, non-cytolytic infection but failed to produce infectious viral particles (Herrel et al., unpublished). ORF4-deficient NYMV therefore has an infection phenotype similar to that of matrix (M) gene-deficient measles virus, Sendai virus (both paramyxoviruses) [10, 26], and rabies virus (a rhabdovirus) [41]. This observation strongly supports the view that ORF4 represents an essential factor for NYMV particle assembly. As the ORF2-derived protein inhibits RdRp activity and further binds to the ORF3-derived protein, the ORF2 product is now speculated to be similar in function to the BDV X protein [23].
NYMV ORF3 expresses a protein that, when expressed together with the ORF1-derived protein (N analog) and the ORF6-derived protein (L analog), reconstitutes full RdRp activity [23]. The ORF3 expression product binds to those of ORF1, ORF2, ORF6, and to itself. These data suggest that ORF3 encodes the analog of mononegavirus polymerase cofactors (P/VP35) [23].
The genome of SbCNV-1 is 11,359 nt long and encodes at least five ORFs (Table 1). ORF1, ORF4, and ORF5 are significantly similar to the N/ORF1, G/ORF5, and L/ORF6 proteins of BDV and MIDWV, respectively. ORF4 encodes a protein with a signal peptide, a carboxy-terminal membrane anchor, and a glycosylation site, suggesting that ORF4 indeed codes for the G analog. ORF2 and ORF3 encode proteins that do not have analogs in common databases, but the predicted ORF2 expression product has a predicted isoelectric point (pI) of 5.1, suggesting that it may be the P/VP35 analog. By the process of elimination and the position of ORF3 in the genome, ORF3 encodes the matrix protein (Table 1) [3].
Life cycle
The cellular life cycles of NYMV, MIDWV, and SbCNV-1 have yet to be characterized. Antisera recognizing the NYMV N/ORF1 and ORF3 expression products strongly stained the nucleus of NYMV-infected cells. Cell fractionation experiments further revealed that NYMV genome-sized RNA is present in the nuclear, but not the cytoplasmic, fraction of infected cells [23]. Nuclear replication is a property NYMV share among the mononegaviruses only with bornaviruses and plant-adapted, arthropod-transmitted nucleorhabdoviruses.
Proposed classification and nomenclature
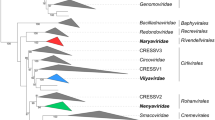
Shotgun sequencing revealed partial NYMV and MIDWV genome fragments with no similarity to other known viruses in the common databases. At the nucleotide level, tBLASTx analysis revealed a distant relationship of some fragments to the RdRps of certain mononegaviruses (bornaviruses, filoviruses, and rhabdoviruses, but not paramyxoviruses) [42]. Phylogenetic analysis of polymerase and nucleoprotein genes by the maximum-parsimony and maximum-likelihood methods revealed that NYMV and MIDWV are indeed most closely related to known mononegaviruses rather than viruses of other taxa, but that NYMV and MIDWV are distantly related to the mononegaviruses that are currently recognized [42] (Fig. 1).
Phylogenetic relationship of NYMV, MIDWV, and SbCNV-1 to other mononegaviruses. A maximum-likelihood tree was constructed using PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/) based on a multiple amino acid sequence alignment of the RdRp polymerase core module as described previously [34]. Virus names and GenBank/RefSeq accession numbers: bornaviruses - Borna disease virus (BDV; NP_042024), avian bornavirus genotype 2 (ABV2; ADU05398); filoviruses - Sudan virus (SUDV; YP_138527), Marburg virus (MARV; YP_001531159); paramyxoviruses - Newcastle disease virus (NDV; NP_071471), Hendra virus (HeV; NP_047113), measles virus (MeV; NP_056924), Sendai virus (SeV; NP_056879), mumps virus (MuV; NP_054714), avian metapneumovirus (AMPV; YP_443845), human respiratory syncytial virus (HRSV; NP_056866); rhabdoviruses - rabies virus (RABV; NP_056797), bovine ephemeral fever virus (BEFV; NP_065409), vesicular stomatitis Indiana virus (VSIV; NP_041716), infectious hematopoietic necrosis virus (IHNV; NP_042681), potato yellow dwarf virus (PYDV; YP_004927971), lettuce necrotic yellows virus (LNYV; YP_425092); nyamiviruses - Nyamanini virus (NYMV; YP_002905337), Midway virus (MIDWV; YP_002905331), soybean cyst nematode virus 1 (SbCNV-1; AEF56729). Obtained and aligned partial or fragment sequences are depicted in boxes. Mononegavirus L-like protein sequences (MLLSs) in pea powdery mildew fungus (Erysiphe pisi) (EpMLLS), yellow fever mosquito (Aedes aegypti) (AaMLLS), zebrafish (Danio rerio) (DrMLLS); nyamivirus L-like protein sequences (NLLSs) in lettuce (Lactuca sativa) (LsNLLS), silkworm (Bombyx mori) (BmNLLS), leafcutter bee (Megachile rotundata) (MrNLLS), black carpenter and fire ants (Camponotus pennsylvanicus and Solenopsis invicta) (CfNLLS and SiNLLS); transcriptome shotgun assembly mononegavirus L-like protein sequences (ShTSA-L) in an ascomycete fungus causing dollar spot (Sclerotinia homoeocarpa) (ShTSA-L). Numbers at the nodes represent aLRT values derived using an SH-like calculation (only values greater than 0.7 are shown). Note that the zebra fish (Animalia: Chordata: Actinopterygii: Cypriniformes: Cyprinidae: Danio rerio Hamilton, 1822) sequence was first reported as another NLLS [4], but is has very poor sequence similarity (≈20 %) to MIDWV L and clusters more with MLLSs from fungi. The sequence is therefore listed as an MLLS (DrMLLS) in the figure
Mihindukulasuriya et al. therefore suggested the establishment of a novel mononegavirus genus, Nyavirus, and discussed the possible need to establish a new mononegavirus family [42]. Like NYMV and MIDWV, SbCNV-1 clearly has many mononegavirus features, but is only very distantly related to known mononegaviruses [3]. All three viruses possess an ORF (ORF1) that encodes a protein with significant similarity to BDV nucleoprotein (N) [42]. Another property shared with bornaviruses (based on experiments with NYMV) is the genomic monophosphorylation at the 5′ end and the protruding 3′ terminus (filoviruses, paramyxoviruses, and rhabdoviruses have triphosphorylated 5′ ends and complementary 5′ and 3′ ends) [23]. NYMV replicates in the host-cell nucleus [23], a feature among the mononegaviruses only shared with nucleorhabdoviruses and bornaviruses. On the other hand, NYMV and possibly also MIDWV, but not SbCNV-1, encode two matrix proteins [23], reminiscent of the coding strategy of filoviruses. In contrast to filoviruses, NYMV, MIDWV, and SbCNV-1 have much shorter genomes (≈11-12 kb compared to ≈18-19 kb in the case of filoviruses) [42], and they form enveloped spherical, rather than filamentous particles [42, 55]. In contrast to most mononegaviruses, but similar to bornaviruses, the polymerase cofactor P of NYMV is encoded by the third rather than the second gene of the viral genome. Like in BDV, the second gene of NYMV codes for a factor that regulates polymerase activity and promotes particle formation [23]. In contrast to bornaviruses, it is unlikely that NYMV uses gene splicing to express additional proteins [23], and in contrast to paramyxoviruses and filoviruses, no ORF contains obvious slippery sequences for cotranscriptional editing. Finally, the NYMV, MIDWV, and SbCNV-1 transcriptional initiation and termination sites are unique [42]. Together, these data clearly indicate that NYMV, MIDWV, and SbCNV-1 are related agents that cannot be assigned to currently existing mononegavirus families (Table 2). Consequently, we propose here the creation of a new family, Nyamiviridae, which includes a single genus, Nyavirus, which in turn includes two separate species for NYMV and MIDWV, and a third, free-floating species for SbCNV-1. Non-Latinized binomial species names distinct from virus names are proposed as recommended in [61] (Table 3), with the exception of the name for the species for SbCNV-1, which may have to be renamed once it is classified into a genus. An official taxonomic proposal (TaxoProp) will be submitted to the ICTV in due time.
Template descriptions of taxa and viruses
Description of Nyamiviridae fam. nov. Kuhn et al ., 2013 (tentative) Etymology of Nyamiviridae: sigil of the first three letters of geo. Nyamanini Pan (place of isolation of Nyamanini virus in South Africa) and the first two letters of geo. Midway Atoll (place of isolation of Midway virus in the USA); and suff. -viridae – ending denoting a virus family [20, 65] → Neo-Lat. n. fem. pl. Nyamiviridae – the family of nyamiviruses.
-
Valid taxon name (fulfills ICVCN Article 3 Rule II-3.8): yes, because name is compliant with ICVCN Article 3 Rules, in particular Rules VI-3.31, VI-3.32, and IX-3.39 [33]; because it has been published [this article]; and because it is associated with descriptive material
-
Accepted name (fulfills ICVCN Article 3 Rule II-3.8): no, because this name has yet to go through the ICTV approval process
-
Use of the taxon:
-
Style: capitalized, italicized, zero article
-
Suggested pronunciation:[ˌnʲɑmɪ’viːrɨdiː] (IPA); nyah-mi-vee-ri-dee (English phonetic notation)
-
Abbreviation: none
-
-
Use of taxon vernaculars:
-
n. sg.: nyamivirus, or, more specific, nyamivirid (-virid: ending denoting a physical member of a virus family [62]). Suggested pronunciation: [ˌnʲɑmɪ’vaɪrəs]/[ˌnʲɑmɪ’viːrɨd] (IPA); nyah-mi-vahy-ruhs/nyah-mi-vee-rid (English phonetic notation)
-
n. pl.: nyamiviruses or, more specific, nyamivirids. Suggested pronunciation:[,nʲɑmɪ’vaɪrəsɨz]/[ˌnʲɑmɪ’viːrɨdz] (IPA); nyah-mi-vahy-ruhs-iz/nyah-mi-vee-ridz (English phonetic notation)
-
adj.: nyamivirus/nyamiviral/nyamivirid. Suggested pronunciation: [,nʲɑmɪ’vaɪrəs]/[ˌnʲɑmɪ’vaɪrəl]/[ˌnʲɑmɪ’viːrɨd] (IPA); nyah-mi-vahy-ruhs/nyah-mi-vahy-ruhl/nyah-mi-vee-rid (English phonetic notation)
-
Style: lower case, not italicized, zero article
-
Abbreviation: none
-
-
Family members share the properties of mononegaviruses [19] and are further characterized by:
-
infecting members of the superphylum Ecdysozoa (in particular arthropods and nematodes) and possibly fungi, cnidarians, and plants, with possible transmission to their hosts (in particular land birds or seabirds in the case of tick-borne viruses)
-
replicating in the nucleus (similar to bornaviruses and nucleorhabdoviruses, but in contrast to paramyxoviruses, filoviruses, and all other rhabdoviruses)
-
having medium-length (≈11 kb) genomes
-
having genomes that are monophosphorylated at the 5′ end and that have protruding 3′ termini (similar to bornaviruses, but in contrast to filoviruses, paramyxoviruses, and rhabdoviruses)
-
having genomes with characteristic transcription initiation and termination signals that are not found in genomes of other mononegaviruses
-
having a lack of dependence on gene splicing for synthesis of replicating progeny
-
having a genomic RNA that encodes 5-6 structural proteins in the order 3′-UTR-N-(VP2)-P-M-G-L-5′-UTR, one of which (VP2) is unique to family members and may correspond to X of bornaviruses and VP24 of filoviruses
-
having an RNA-dependent RNA polymerase, whose core domain sequence separates them clearly from other mononegaviruses
-
maturing by envelopment of independently assembled nucleocapsids at membrane sites containing inserted virus proteins; budding occurs predominantly from the plasma membrane; the virion envelope is derived from the host-cell membrane and is considered to have a lipid composition similar to that of the plasma membrane
-
forming spherical virions
-
forming virions that are ≈100–130 nm in diameter
-
-
Suggested type genus: Nyavirus (the ICTV currently does not endorse the status of type genus)
-
Included genera: genus Nyavirus (tentative, see below)
Description of Nyavirus gen. nov. Mihindukulasuriya et al ., 2009 [ 42 ] (tentative) Etymology of Nyavirus: sigil of the first three letters of geo. Nyamanini Pan (place of isolation of Nyamanini virus in South Africa); and -virus – ending denoting a virus genus [65] Neo-Lat. n. neut. sg. Nyavirus – the genus of nyaviruses.
-
Valid taxon name (fulfills ICVCN Article 3 Rule II-3.8): yes, because name is compliant with ICVCN Article 3 Rules, in particular Rules IV-3.26, IV-3.27, IV-3.28, and IX-3.39 [33]; because it has been published [42]; and because it is associated with descriptive material
-
Accepted name (fulfills ICVCN Article 3 Rule II-3.8): no, because this name has yet to go through the ICTV approval process
-
Use of the taxon:
-
Style: capitalized, italicized, zero article
-
Suggested pronunciation: [ˌnʲɑ’vaɪrəs] (IPA); nyah-vahy-ruhs (English phonetic notation)
-
Abbreviation: none
-
-
Use of taxon vernaculars:
-
n. sg.: nyavirus (-virus: ending denoting a physical member of a virus genus [62]). Suggested pronunciation: [ˌnʲɑ’vaɪrəs] (IPA); nyah-vahy-ruhs (English phonetic notation)
-
n. pl. nyaviruses. Suggested pronunciation: [ˌnʲɑ’vaɪrəsɨz] (IPA); nyah-vahy-ruhs-iz (English phonetic notation)
-
adj.: nyavirus/nyaviral. Suggested pronunciation: [ˌnʲɑ’vaɪrəs]/[ˌnʲɑ’vaɪrəl] (IPA); nyah-vahy-ruhs/nyah-vahy-ruhl (English phonetic notation)
-
Style: lower case, not italicized, one word, zero article
-
Abbreviation: none
-
-
Genus members are characterized by having the properties of nyamiviruses (because there is currently only one nyamivirus genus)
-
Type species: Nyamanini nyavirus (tentative, see below)
-
Included species: species Nyamanini nyavirus (tentative, see below), Midway nyavirus (tentative, see below)
Description of Nyamanini nyavirus sp. nov. Kuhn et al ., 2013 (tentative) Etymology of Nyamanini nyavirus: derived from geo. Nyamanini Pan (place of isolation of Nyamanini virus in South Africa) → the Nyamanini species of nyamiviruses.
-
Valid taxon name (fulfills ICVCN Article 3 Rule II-3.8): yes, because name is compliant with ICVCN Article 3 Rules, in particular Rule IX-3.40 [33]; because it has been published [this article]; and because it is associated with descriptive material
-
Accepted name (fulfills ICVCN Article 3 Rule II-3.8): no, because this name has yet to go through the ICTV approval process
-
Use of the taxon:
-
Style: capitalized, italicized, zero article
-
Suggested pronunciation: [ˌnʲɑ’mɑnɪnɪ ˌnʲɑ’vaɪrəs] (IPA); nyah-mah-ni-ni nyah-vahy-ruhs (English phonetic notation)
-
Abbreviation: none
-
-
Use of taxon vernaculars:
-
n. sg.: Nyamanini nyavirus. Suggested pronunciation: [ˌnʲɑ’mɑnɪnɪ ˌnʲɑ’vaɪrəs] (IPA); nyah-mah-ni-ni nyah-vahy-ruhs (English phonetic notation)
-
n. pl. Nyamanini nyaviruses. Suggested pronunciation: [ˌnʲɑ’mɑnɪnɪ ˌnʲɑ’vaɪrəsɨz] (IPA); nyah-mah-ni-ni nyah-vahy-ruhs-iz (English phonetic notation)
-
adj.: Nyamanini nyavirus/Nyamanini nyaviral. Suggested pronunciation: [ˌnʲɑ’mɑnɪnɪ ˌnʲɑ’vaɪrəs]/[ˌnʲɑ’mɑnɪnɪ ˌnʲɑ’vaɪrəl] (IPA); nyah-mah-ni-ni nyah-vahy-ruhs/nyah-mah-ni-ni nyah-vahy-ruhl (English phonetic notation)
-
Style: lower case, not italicized, one word, zero article
-
Abbreviation: none
-
Species members are characterized by having the properties of nyaviruses; having a full-length genomic sequence different from the type virus of the type species of the genus Nyavirus (Nyamanini virus) by <30 %; and infecting soft ticks and/or land birds
-
-
Suggested type virus: Nyamanini virus (NYMV)
-
Species members: Nyamanini virus (NYMV)
Description of Midway nyavirus sp. nov. Kuhn et al ., 2013 (tentative) Etymology of Midway nyavirus: derived from geo. Midway Atoll (place of isolation of Midway virus in the USA); and Nyavirus – the genus of nyaviruses → the Midway species of nyaviruses.
-
Valid taxon name (fulfills ICVCN Article 3 Rule II-3.8): yes, because name is compliant with ICVCN Article 3 Rules, in particular Rule IX-3.40 [33]; because it has been published [this article]; and because it is associated with descriptive material
-
Accepted name (fulfills ICVCN Article 3 Rule II-3.8): no, because this name has yet to go through the ICTV approval process
-
Use of the taxon:
-
Style: capitalized, italicized, zero article
-
Suggested pronunciation: [ˈmɪdˌweɪ ˌnʲɑ’vaɪrəs] (IPA); mid-wey nyah-vahy-ruhs (English phonetic notation)
-
Abbreviation: none
-
-
Use of taxon vernaculars:
-
n. sg.: Midway nyavirus. Suggested pronunciation: [ˈmɪdˌweɪ ˌnʲɑ’vaɪrəs] (IPA); mid-wey nyah-vahy-ruhs (English phonetic notation)
-
n. pl. Midway nyaviruses. Suggested pronunciation: [ˈmɪdˌweɪ ˌnʲɑ’vaɪrəsɨz] (IPA); mid-wey nyah-vahy-ruhs-iz (English phonetic notation)
-
adj.: Midway nyavirus/Midway nyaviral. Suggested pronunciation: [ˈmɪdˌweɪ ˌnʲɑ’vaɪrəs]/[ˈmɪdˌweɪˌnʲɑ’vaɪrəl] (IPA); mid-wey nyah-vahy-ruhs/mid-wey nyah-vahy-ruhl (English phonetic notation)
-
Style: lower case, not italicized, one word, zero article
-
Abbreviation: none
-
Species members are characterized by having the properties of nyaviruses; having a full-length genomic sequence different from the type virus of the type species of the genus Nyavirus (Nyamanini virus) by >30 % and that of Midway virus <30 %; and infecting soft ticks and seabirds
-
-
Suggested type virus: Midway virus (MIDWV)
-
Species members: Midway virus (MIDWV)
Description of Soybean cyst nematode virus sp. nov. Kuhn et al ., 2013 (tentative) Etymology of Soybean cyst nematode virus: derived from the host of the virus (the soybean cyst nematode (Heterodera glycines)) → the soybean cyst nematode species of nyamiviruses.
-
Valid taxon name (fulfills ICVCN Article 3 Rule II-3.8): yes, because name is compliant with ICVCN Article 3 Rules, in particular Rule IX-3.40 [33]; because it has been published [this article]; and because it is associated with descriptive material
-
Accepted name (fulfills ICVCN Article 3 Rule II-3.8): no, because this name has yet to go through the ICTV approval process
-
Use of the taxon:
-
Style: capitalized, italicized, zero article
-
Suggested pronunciation: [ˈsɔɪˌbin sɪst ˈnɛməˌtoʊd ‘vaɪrəs] (IPA); soi-been sistnem-uh-tohd vahy-ruhs (English phonetic notation)
-
Abbreviation: none
-
-
Use of taxon vernaculars:
-
n. sg.: soybean cyst nematode virus. Suggested pronunciation: [ˈsɔɪˌbin sɪst ˈnɛməˌtoʊd ‘vaɪrəs] (IPA); soi-been sist nem-uh-tohd vahy-ruhs (English phonetic notation)
-
n. pl. soybean cyst nematode viruses. Suggested pronunciation: [ˈsɔɪˌbin sɪst ˈnɛməˌtoʊd vaɪrəsɨz] (IPA); soi-been sist nem-uh-tohd vahy-ruhs-iz (English phonetic notation)
-
adj.: soybean cyst nematode virus/soybean cyst nematode viral. Suggested pronunciation: [ˈsɔɪˌbin sɪst ˈnɛməˌtoʊd ˈvaɪrəs]/[ˈsɔɪˌbin sɪst ˈnɛməˌtoʊd ˈvaɪrəl] (IPA); soi-been sist nem-uh-tohd vahy-ruhs/soi-been sist nem-uh-tohd vahy-ruhl (English phonetic notation)
-
Style: lower case, not italicized, one word, zero article
-
Abbreviation: none
-
-
Species members are characterized by having the properties of nyamiviruses; having a full-length genomic sequence different from the type virus of the type species of the genus Nyavirus (Nyamanini virus) by >30 % and that of soybean cyst nematode virus 1 <30 %, and infecting nematodes
-
Suggested type virus: soybean cyst nematode virus 1 (SbCNV-1)
-
Species members: soybean cyst nematode virus 1 (SbCNV-1)
Description of Nyamanini virus vir. McIntosh, 1960 (as described in [ 57 ]) Etymology of Nyamanini virus: derived from geo. Nyamanini Pan (place of isolation of Nyamanini virus in South Africa (the word is probably derived from from Zulu *inyamanini (place where there is little meat) and is composed of <inyama (meat) + -ani- (diminutive suffix) + -ini- (locative suffix)); and Lat. n. neut. sg. virus – poison, slime, venom.
-
Use of the name:
-
Style: first word capitalized (because proper noun), not italicized, all types of articles
-
Suggested pronunciation: [ˌnʲɑ’mɑnɪnɪ ‘vaɪrəs] (IPA); nyah-mah-ni-ni vahy-ruhs (English phonetic notation)
-
Abbreviation: NYMV
-
-
Virus variants are characterized by having the properties of Nyamanini nyaviruses plus:
-
diverging in genomic nucleotide sequence from the type variant of the type virus of the species Nyamanini nyavirus (Wat-tick39) by <10 %
-
-
Suggested type variant: Wat-tick39
Description of Midway virus vir. Yunker, 1975 [ 68 ] Etymology of Midway virus: derived from geo. Midway Atoll (place of isolation of Midway virus in the USA); and Lat. n. neut. sg. virus – poison, slime, venom.
-
Use of the name:
-
Style: first word capitalized (because proper noun), not italicized, all types of articles
-
Suggested pronunciation: [ˈmɪdˌweɪ ‘vaɪrəs] (IPA); mid-wey vahy-ruhs (English phonetic notation)
-
Abbreviation: MIDWV
-
-
Virus variants are characterized by having the properties of Midway nyaviruses plus:
-
diverging in genomic nucleotide sequence from the type variant of the type virus of the species Midway nyavirus (Mid-RML47153) by <10 %
-
-
Suggested type variant: Mid-RML47153
Description of soybean cyst nematode virus 1 vir. Bekal et al ., 2011 [ 3 ] emend. Kuhn et al ., 2013 Etymology of soybean cyst nematode virus 1: derived from the host of the virus (the soybean cyst nematode); and Lat. n. neut. sg. virus – poison, slime, venom.
-
Use of the name:
-
Style: lower case, not italicized, all types of articles
-
Suggested pronunciation: [ˈsɔɪˌbin sɪst ˈnɛməˌtoʊd ‘vaɪrəs] (IPA); soi-been sist nem-uh-tohd vahy-ruhs (English phonetic notation)
-
Abbreviation: SbCNV-1
-
-
Virus variants are characterized by having the properties of soybean cyst nematode viruses 1 plus:
-
diverging in genomic nucleotide sequence from the type variant of the type virus of the species Soybean cyst nematode virus (Urb-10) by <10 %
-
-
Suggested type variant: Urb-10
Proposal for nomenclature below the species level
We propose to adapt the recently published nomenclature scheme for filovirus [35] variants for nyamivirus variants. Accordingly, nyamiviruses would be named <virus name>/<isolation host-suffix>/<country of sampling>/<year of sampling>/<genetic variant designation>-<isolate designation>. Instructions on how to fill the individual fields can be found in [35]. Furthermore, we strongly suggest not using the word “strain” in context with nyamiviruses for reasons also explained in detail in [35]. A list of nyamivirus variants, named according to the scheme presented in [35] can be found in Table 4.
Proposal for preliminary gene and protein designations
We propose to follow general mononegavirus gene and protein nomenclature in regard to nyamiviruses. Accordingly, ORF1 of NYMV, MIDWV, and SbCNV-1 would be part of the N gene, encoding the nucleoprotein (N). ORF3 of NYMV and MIDWV and ORF2 of SbCNV-1 would be part of the P gene and encode the phosphoprotein (P). ORF4 of NYMV and MIDWV and ORF3 of SbCNV-1 would be part of the M gene and encode the major matrix protein (M). ORF5 of NYMV and MIDWV and ORF4 of SbCNV-1 would be part of the G gene and encode the glycoprotein (G). Finally, ORF6 of NYMV and MIDWV and ORF5 of SbCNV-1 would be part of the L gene and encode the RNA-dependent RNA polymerase (L). As the expression product of ORF2 of NYMV and MIDWV has to be further characterized to assign function, we propose to assign only provisional names to the ORF2 gene (“VP2”) and the expressed protein (VP2).
Future prospects
The proposed family Nyamiviridae will most likely have to be expanded in the future. For instance, in 1975 a virus, provisionally called “BA-T” (bovine abortion – tick), was isolated from ticks (Animalia: Arthropoda: Arachnida: Ixodida: Argasidae: Ornithodoros coriaeous Koch, 1844) collected in northern California during an outbreak of bovine abortion due to bacterial infection. The recent determination of BA-T’s full genomic sequence revealed clear homology with NYMV and MIDWV, although it proved to be antigenically distinct, and although it produces larger virions (RBT unpublished). The discovery of SbCNV-1 in the TN10 inbred line of the soybean cyst nematode suggests that nematodes in general may carry similar viruses. Indeed, a recent conference abstract suggests that SbCNV-1 is also present in North Carolina race 4 (NC4) of this nematode [51], and a nucleic acid fragment very similar to SbCNV-1 was found in a sugar beet nematode culture (Animalia: Nematoda: Chromadorea: Tylenchida: Heteroderidae: Heterodera schachtii Schmidt, 1871) [ 3 ]. Likewise, the virome of ticks is under-studied, and it hardly would be a surprise if NYMV- or MIDWV-related viruses were discovered in various ticks or their hosts.
Kondo et al., recently detected mononegavirus L-like protein sequences (MLLSs) in the genome of the pea powdery mildew fungus (Fungi: Ascomyctoa: Leotomycetes: Erysiphales: Erysiphaceae: Erysiphe pisi DC., 1821), as well as in transcriptome shotgun assembly (TSA) libraries established from another fungus (Fungi: Ascomycota: Ascomycetes: Helotiales: Sclerotiniaceae: Sclerotina homeocarpa Benn, 1937) (Fig. 1) [34]. Nyamivirus L-like protein sequences (NLLSs) were then detected in the genomes of silkworms (Animalia: Arthropoda: Insecta: Lepidoptera: Bombycidae: Bombyx mori Linnaeus, 1758), black carpenter ants (Insecta: Hymenoptera: Formicidae: Camponotus pennsylvanicus de Geer, 1773), fire ants (Insecta: Hymenoptera: Formicidae: Solenopsis invicta Buren, 1972), leafcutter bees (Insecta: Hymenoptera: Megachilidae: Megachile (Eutricharaea) rotundata Fabricius 1787) and lettuce (Plantae: Asterales: Asteraceae: Lactuca sativa Linnaeus, 1753) (Fig. 1) [34]. A nyamivirus L-like protein sequence (similarity to NYMV L 28 %) was also detected in the pavement ant (Animalia: Arthropoda: Insecta: Hymenoptera: Formicidae: Myrmicinae: Tetramorium caespitum Linnaeus, 1758) and mentioned in a dissertation. The sequence of this virus was elongated to a total of 12,095 nt, but has not yet been made available for analysis [52]. Preliminary database searches also led to the detection of several nyamivirus N(P)- and G(P)-like protein sequences in genomic material associated with hymenoptera (fire ants and leafcutter bees), soybean cyst nematodes, pine wilt nematodes (Animalia: Nematoda: Secernentea: Aphelenchida: Parasitaphelenchidae: Bursaphelenchus xylophilus Steiner & Buhrer, 1934), northern root-knot nematodes (Animalia: Nematoda: Secernentea: Heteroderidae: Meloidogyne hapla Chitwood, 1949), southern root-knot nematodes (Animalia: Nematoda: Secernentea: Heteroderidae: Meloidogyne incognita Kofoid & White, 1919), nematodes that do not yet have a vernacular name (Animalia: Nematoda: Secernentea: Rhabditida: Caenorhabditis angaria Sudhaus et al., 2011), and the warty comb jelly (Animalia: Ctenophora: Bolinopsidae: Mnemiopsis leidyi Agassiz, 1865) (HK and NS unpublished results).
Importantly, we do not propose that any of these short nucleic acid fragments should be taxonomically classified. The determination of nearly complete genome sequences and determination of the ecological niche in which these genomes can be detected is the minimal set of data we propose is necessary for taxonomic consideration. However, the detected NLLSs clearly suggest that nyamiviruses may have a very wide host distribution and that many new nyamiviruses may soon be discovered.
Abbreviations
- adj :
-
adiectivum (adjective)
- fam. nov :
-
familia nova (new family)
- fem :
-
femininum
- gen. nov :
-
genus novum (new genus)
- geo :
-
geographic
- IPA :
-
International Phonetic Alphabet
- Lat :
-
Latin
- n :
-
nomen substantivum (noun)
- neo-Lat :
-
Neo-Latin
- neut :
-
neutrum
- pl :
-
numerus pluralis (plural)
- sg :
-
numerus singularis (singular)
- sp. nov :
-
species nova (new species)
- suff :
-
suffix
- vir :
-
virus
References
Adams MJ, Christian P, Ghabrial SA, Knowles NJ, Lavigne R (2011) Unassigned Viruses. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy—Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, pp 1199–1207
Adams MJ, Carstens EB (2012) Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012). Arch Virol 157:1411–1422
Bekal S, Domier LL, Niblack TL, Lambert KN (2011) Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J Gen Virol 92:1870–1879
Belyi VA, Levine AJ, Skalka AM (2010) Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Path 6:e1001030
Berge TO (1975) International catalogue of arboviruses; including certain other viruses of vertebrates, 2nd edn. US Department of Health, Education, and Welfare, Public Health Service, Center for Disease Control, Atlanta
Bergh O, Borsheim KY, Bratbak G, Heldal M (1989) High abundance of viruses found in aquatic environments. Nature 340:467–468
Bhat UKM, Yunker CE (1979) Susceptibility of a tick cell line (Dermacentor parumapertus Neumann) to infection with arboviruses. In: Kurstak E (ed) Arctic and tropical arboviruses. Academic Press, New York, pp 263–275
Calisher CH, Carstens EB, Christian P, Mahy BWJ, Mayo MA, Shope RE (2000) Unassigned Viruses. In: van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB (eds) Virus taxonomy—Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, pp 995–1008
Casals J (1968) Filtration of arboviruses through ‘Millipore’ membranes. Nature 217:648–649
Cathomen T, Mrkic B, Spehner D, Drillien R, Naef R, Pavlovic J, Aguzzi A, Billeter MA, Cattaneo R (1998) A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J 17:3899–3908
Cerny V (1983) Las aves infectadas con garrapatas Ornitodorus vectoras de virus. Rev Cub Med Trop 35:181–185 (Spanish)
Chastel C (1980) Arbovirus transmis par des tiques et associés à des oiseaux de mer; une revue générale. Méd Trop 40:535–548 (French)
Clifford CM, Thomas LA, Hughes LE, Kohls GM, Philip CB (1968) Identification and comparison of two viruses isolated from ticks of the genus Ornithodoros. Am J Trop Med Hyg 17:881–885
Clifford CM (1979) Tick-borne viruses of seabirds. In: Kurstak E (ed) Arctic and tropical arboviruses. Academic Press, New York, pp 83–100
Culley AI, Lang AS, Suttle CA (2007) The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol J 4:69
Darwish MA, Imam IZE, Omar FM, el-Karamany RM (1975) Antibodies to certain tick-borne viruses in Egyptian sera. J Egypt Publ Health Assoc 50:37–42
Djikeng A, Kuzmickas R, Anderson NG, Spiro DJ (2009) Metagenomic analysis of RNA viruses in a fresh water lake. PLoS ONE 4:e7264
Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Rasche A, Yordanov S, Seebens A, Oppong S, Adu Sarkodie Y, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stocker A, Carneiro AJ, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EK, Kruppa T, Franke CR, Kallies R, Yandoko ER, Herrler G, Reusken C, Hassanin A, Kruger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C (2012) Bats host major mammalian paramyxoviruses. Nat Commun 3:796
Easton AJ, Pringle CR (2011) Order Mononegavirales. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy—Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, pp 653–657
Fenner F (1976) Classification and nomenclature of viruses—Second Report of the International Committee on Taxonomy of Viruses. Intervirology 7:1–115
Francki RIB, Fauquet CM, Knudson DL, Brown F (1991) Classification and nomenclature of viruses—Fifth Report of the International Committee on Taxonomy of Viruses. Archives of Virology Supplement, vol. 2. Springer, Vienna
Grard G, Fair JN, Lee D, Slikas E, Steffen I, Muyembe JJ, Sittler T, Veeraraghavan N, Ruby JG, Wang C, Makuwa M, Mulembakani P, Tesh RB, Mazet J, Rimoin AW, Taylor T, Schneider BS, Simmons G, Delwart E, Wolfe ND, Chiu CY, Leroy EM (2012) A novel rhabdovirus associated with acute hemorrhagic fever in central Africa. PLoS Path 8:e1002924
Herrel M, Hoefs N, Staeheli P, Schneider U (2012) Tick-borne Nyamanini virus replicates in the nucleus and exhibits unusual genome and matrix protein properties. J Virol 86:10739–10747
Hoogstraal H (1973) Viruses and ticks. In: Gibbs AJ (ed) Viruses and invertebrates. North Holland Publishing Co., Amsterdam, pp 349–390
Hoogstraal H, Kaiser MN, McClure HE (1974) The subgenus Persicargas (Ixodoidea: Argasidae: Argas) 20. A. (P.) robertsi parastizing nesting wading birds and domestic chickens in the Australian and oriental regions, viral infections, and host migration. J Med Entomol 11:513–524
Inoue M, Tokusumi Y, Ban H, Kanaya T, Shirakura M, Tokusumi T, Hirata T, Nagai Y, Iida A, Hasegawa M (2003) A new Sendai virus vector deficient in the matrix gene does not form virus particles and shows extensive cell-to-cell spreading. J Virol 77:6419–6429
Jupp PG, McIntosh BM (1981) Ticks and arthropod-borne viruses in South Africa. Tick biology and control. In: Proceedings of an International Conference held from 27–29 January 1981, under the auspices of the Tick Research Unit, Rhodes University, Grahamstown, South Africa, pp 177–180
Jupp PG, McIntosh BM (1986) Identity of argasid ticks yielding isolations of Chenuda, Quaranfil and Nyamanini viruses in South-Africa. J Entomol Soc South Afr 49:392
Kaiser MN (1966) Viruses in ticks. I. Natural infections of Argas (Persicargas) arboreus by Quaranfil and Nyamanini viruses and absence of infections in A. (P.) persicus in Egypt. Am J Trop Med Hyg 15:964–975
Karabatsos N (1985) International catalogue of arboviruses, including certain other viruses of vertebrates, 3rd edn. American Society of Tropical Medicine and Hygiene for The Subcommittee on Information Exchange of the American Committee on Arthropod-borne Viruses, San Antonio
Kemp GE, Lee VH, Moore DL (1975) Isolation of Nyamanini and Quaranfil viruses from Argas (Persicargas) arboreus ticks in Nigeria. J Med Entomol 12:535–537
King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) (2011) Virus taxonomy—Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London
King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (2011) The International Code of Virus Classification and Nomenclature. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy–Ninth Report of the International Committee on Taxonomy of viruses. Elsevier/Academic Press, London, pp 1273–1277
Kondo H, Chiba S, Toyoda K, Suzuki N (2013) Evidence for negative-strand RNA virus infection in fungi. Virology 435:201–209
Kuhn JH, Bao Y, Bavari S, Becker S, Bradfute S, Brister JR, Bukreyev AA, Chandran K, Davey RA, Dolnik O, Dye JM, Enterlein S, Hensley LE, Honko AN, Jahrling PB, Johnson KM, Kobinger G, Leroy EM, Lever MS, Muhlberger E, Netesov SV, Olinger GG, Palacios G, Patterson JL, Paweska JT, Pitt L, Radoshitzky SR, Saphire EO, Smither SJ, Swanepoel R, Towner JS, van der Groen G, Volchkov VE, Wahl-Jensen V, Warren TK, Weidmann M, Nichol ST (2013) Virus nomenclature below the species level: a standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch Virol 158:301–311
Li L, Delwart E (2011) From orphan virus to pathogen: the path to the clinical lab. Curr Opin Virol 1:282–288
Matthews REF (1979) Classification and nomenclature of viruses—Third Report of the International Committee on Taxonomy of Viruses. Report prepared by the International Committee on Taxonomy of Viruses and published for the Virology Section of the International Association of Microbiological Societies (IAMS). Intervirology 12:129–296
Matthews REF (1982) Classification and nomenclature of viruses—Fourth Report of the International Committee on Taxonomy of viruses. Report prepared by the International Committee on Taxonomy of viruses and published for the Virology Division of the International Union of Microbiological Societies (IUMS). Intervirology 17:1–199
Mayo MA (1995) Unassigned viruses. In: Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD (eds) Virus taxonomy—Sixth Report of the International Committee on Taxonomy of Viruses Archives of Virology Supplement, vol 10., Springer, Vienna, Austria, pp 504–507
Mayo MA, Christian PD, Hillmann BI, Burunt AA, Desselberger U (2005) Unassigned viruses. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy—Eighth Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, pp 1131–1144
Mebatsion T, Weiland F, Conzelmann KK (1999) Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J Virol 73:242–250
Mihindukulasuriya KA, Nguyen NL, Wu G, Huang HV, da Rosa AP, Popov VL, Tesh RB, Wang D (2009) Nyamanini and Midway viruses define a novel taxon of RNA viruses in the order Mononegavirales. J Virol 83:5109–5116
Mohammed YS, Grešíková M, Adamyová K, Ragib AH, el-Dawala K (1970) Studies on arboviruses in Egypt. II. Contribution of arboviruses to the aetiology of undiagnosed fever among children. J Hyg 68:491–495
Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD (1995) Virus taxonomy—Sixth Report of the International Committee on Taxonomy of Viruses. Archives of Virology Supplement, vol. 10. Springer, Vienna
Negredo A, Palacios G, Vazquez-Moron S, Gonzalez F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, de la Cruz Martinez M, Herrera JE, Pizarro M, Hutchison SK, Echevarria JE, Lipkin WI, Tenorio A (2011) Discovery of an ebolavirus-like filovirus in Europe. PLoS Path 7:e1002304
Pringle CR, Alexander DJ, Billeter MA, Collins PL, Kingsbury DW, Lipkind MA, Nagai Y, Orvell C, Rima B, Rott R, ter Meulen V (1991) The order Mononegavirales. Arch Virol 117:137–140
Pringle CR (1997) The Order Mononegavirales—current status. Arch Virol 142:2321–2326
Rosario K, Duffy S, Breitbart M (2009) Diverse circovirus-like genome architectures revealed by environmental metagenomics. J Gen Virol 90:2418–2424
Rosario K, Nilsson C, Lim YW, Ruan Y, Breitbart M (2009) Metagenomic analysis of viruses in reclaimed water. Environ Microbiol 11:2806–2820
Rosario K, Breitbart M (2011) Exploring the viral world through metagenomics. Curr Opin Virol 1:289–297
Ruark CL, Lommel S (2012) Nematodes: a suitable home for viruses? A potential opportunity for biocontrol. In: Abstracts of the 11th Annual Summer Undergraduate Research Symposium, August 1, North Carolina State University, Raleigh, North Carolina, USA, pp 75 (session 72, D73)
Runckel C (2012) Determinants of recombination in RNA viruses. PhD dissertation in biochemistry. University of California, San Francisco
Skennerton CT, Angly FE, Breitbart M, Bragg L, He S, McMahon KD, Hugenholtz P, Tyson GW (2011) Phage encoded H-NS: a potential achilles heel in the bacterial defence system. PLoS ONE 6:e20095
Takahashi M, Nii S, Casals J (1980) Hirota virus: a new tick-borne arbovirus related to Nyamanini. In: Vesenjak-Hirjan J (ed) Arboviruses in the Mediterranean countries: 6th FEMS symposium held under the auspices of the Yugoslav Academy of Sciences and Arts in Zagreb. Zentralblatt für Bakteriologie, Mikrobiologie und Hygiene - 1. Abteilung Supplement 9. Gustav Fischer Verlag, Stuttgart
Takahashi M, Yunker CE, Clifford CM, Nakano W, Fujino N, Tanifuji K, Thomas LA (1982) Isolation and characterization of Midway virus: a new tick-borne virus related to Nyamanini. J Med Virol 10:181–193
Taylor RM, Henderson JR, Thomas LA (1966) Antigenic and other characteristics of Quaranfil, Chenuda, and Nyamanini arboviruses. Am J Trop Med Hyg 15:87–90
Taylor RM, Hurlbut HS, Work TH, Kingston JR, Hoogstraal H (1966) Arboviruses isolated from Argas ticks in Egypt: Quaranfil, Chenuda, and Nyamanini. Am J Trop Med Hyg 15:76–86
Taylor RM (1967) Catalogue of arthropod-borne viruses of the world: a collection of data on registered arthropod-borne animal viruses. US Public Health Service, Washington, DC
Theiler M, Downs WG (1973) The arthropod-borne viruses of vertebrates. An account of the Rockefeller Foundation Virus Program 1951–1970. Yale University Press, New Haven
Tucker KP, Parsons R, Symonds EM, Breitbart M (2011) Diversity and distribution of single-stranded DNA phages in the North Atlantic Ocean. ISME J 5:822–830
Van Regenmortel MH, Burke DS, Calisher CH, Dietzgen RG, Fauquet CM, Ghabrial SA, Jahrling PB, Johnson KM, Holbrook MR, Horzinek MC, Keil GM, Kuhn JH, Mahy BW, Martelli GP, Pringle C, Rybicki EP, Skern T, Tesh RB, Wahl-Jensen V, Walker PJ, Weaver SC (2010) A proposal to change existing virus species names to non-Latinized binomials. Arch Virol 155:1909–1919
Vetten HJ, Haenni AL (2006) Taxon-specific suffixes for vernacular names. Arch Virol 151:1249–1250
Walker JB (1977) Ticks and human disease in tropical Africa. Medicine in a tropical environment. In: Proceedings of the International Symposium, South Africa/1976, pp 276–290
Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95:6578–6583
Wildy P (1971) Classification and nomenclature of viruses—First Report of the International Committee on Nomenclature of Viruses. S. Karger, Basel
Yin Y, Fischer D (2008) Identification and investigation of ORFans in the viral world. BMC Genomics 9:24
Yunker CE (1970) Worldwide research on human and animal diseases caused by tickborne viruses. Misc Publ Entomol Soc Am 6:330–338
Yunker CE (1975) Tick-borne viruses associated with seabirds in North America and related islands. Med Biol 53:302–311
Zeller HG, Mitchell CJ (1989) Replication of certain recently classified viruses in Toxorhynchites amboinensis mosquitoes and mosquito and mammalian cell lines, with implications for their arthropod-borne status. Res Virol 140:563–570
Acknowledgments
We thank Robert Swanepoel (Zoonoses Research Unit, University of Pretoria, Pretoria, South Africa) for clarification regarding the pronunciation of the word “Nyamanini,” and Laura Bollinger (Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Fort Detrick, Frederick, Maryland, USA) for critically reviewing and editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclaimer
The taxonomic changes suggested here have not been endorsed by the International Committee on Taxonomy of Viruses (ICTV) Executive Committee, may differ from any new taxonomy that is ultimately approved by the ICTV, and is presented for discussion only but has no official standing at the time of publication. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, the US Department of Defense, the US Department of the Army or of the institutions and companies affiliated with the authors. YC and JHK performed this work as employees of Tunnell Consulting, Inc., and ANC performed this work as the owner of Logos Consulting, Inc. Both companies are subcontractors to Battelle Memorial Institute under its prime contract with NIAID, under Contract No. HHSN272200200016I.
Jens H. Kuhn, Ralf G. Dietzgen, Norbert Nowotny, and Lin-Fa Wang are the members of the International Committee on Taxonomy of Viruses (ICTV) Mononegavirales Study Group, and are the chairs of the ICTV Filoviridae, Rhabdoviridae, Bornaviridae, and Paramyxoviridae Study Groups, respectively.
Rights and permissions
About this article
Cite this article
Kuhn, J.H., Bekal, S., Caì, Y. et al. Nyamiviridae: Proposal for a new family in the order Mononegavirales . Arch Virol 158, 2209–2226 (2013). https://doi.org/10.1007/s00705-013-1674-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1674-y