Abstract.
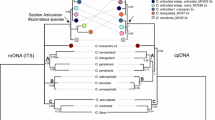
A phylogenetic study of the largest tribe of palms, the Areceae, was conducted using sequences of two low-copy nuclear genes. Previous morphological and plastid DNA studies have not supported the monophyly of the tribe, but have placed its members in a large clade that includes the monophyletic tribes Geonomeae, Cocoeae, Podococceae, and Hyophorbeae. We analyzed this large clade to test the monophyly of tribe Areceae with nuclear data, to explore relationships among its subtribes, and to identify other monophyletic groups. For 54 palm species, including members of all 17 subtribes of tribe Areceae, we sequenced regions of the malate synthase (MS) and phosphoribulokinase (PRK) genes. Simultaneous analysis of these regions revealed 52 shortest trees, all of which resolved tribe Areceae as polyphyletic. Subtribes Iguanurinae, Dypsidinae, Oncospermatinae, and Arecinae were also resolved as polyphyletic. A clade of Indo-Pacific taxa was resolved with strong support, and would be a suitable target for more focused study.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received February 7, 2001; accepted April 9, 2002 Published online: December 3, 2002
Rights and permissions
About this article
Cite this article
Lewis, C., Doyle, J. A phylogenetic analysis of tribe Areceae (Arecaceae) using two low-copy nuclear genes. Plant Syst. Evol. 236, 1–17 (2002). https://doi.org/10.1007/s00606-002-0205-1
Issue Date:
DOI: https://doi.org/10.1007/s00606-002-0205-1