Abstract
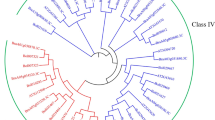
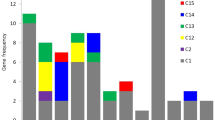
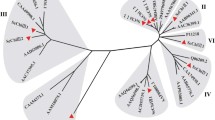
Chitinolytic enzymes are important pathogenesis and stress related proteins. We identified 27 putative genes encoding endochitinases in the maize genome via in silico techniques and four exochitinases. Only seven of the endochitinases and segments of the exochitinases were heretofore known. The endochitinases included members of family 19 chitinases (classes I–IV of PR3, II of PR4) and members of family 18 chitinases (class III of PR8). Some similar enzymes were detected on adjacent regions of the same chromosome, and seem to result from duplication events. Most of the genes expressed were identified from EST libraries from plants exposed to biotic or abiotic stresses but also from libraries from tissues not exposed to stresses. We isolated proteins from seedlings of maize in the presence or absence of the symbiotic root colonizing fungus Trichoderma harzianum strain T22, and analyzed the activity of chitinolytic enzymes using an in-gel activity assay. The activity bands were identified by LC/MS/MS using the database from our in silico study. The identities of the enzymes changed depending on whether or not T22 was present. One activity band of about 95 kDa appeared to be a heterodimer between an exochitinase and any of several different endochitinases. The identity of the endochitinase component appeared to be dependent upon treatment.








Similar content being viewed by others
References
Baldan B, Guzzo F, Filippini F, Gasparian M, LoSchiavo F, Vitale A, de Vries SC, Mariani P, Terzi M (1997) The secretory nature of the lesion of carrot cell variant ts11, rescuable by endochitinase. Planta 203:381–389
Berger S, Menudier A, Julien R, Karamanos Y (1995) Do de-N-glycosylation enzymes have an important role in plant cells? Biochimie 77:751–760
Bolar JP, Norelli JL, Wong K-W, Hayes CK, Harman GE, Aldwinckle HS (2000) Expression of endochitinase from Trichoderma harzianum in transgenic apple increases resistance to apple scab and reduces vigor. Phytopathology 90:72–77
Bolar JP, Norelli JL, Harman GE, Brown SK, Aldwinckle HS (2001) Synergistic activity of endochitinase and exochitinase from Trichoderma atroviride (T. harzianum) against the pathogenic fungus (Venturia inaequalis) in transgenic apple plants. Trans Res 10:533–543
Bravo JM, Campo S, Murillo I, Coca M, San Segundo B (2003) Fungus- and wound-induced accumulation of mRNA containing a class II chitinase of the pathogenesis-related protein 4 (PR-4) family of maize. Plant Mol Biol 52:745–759
Chevalier C, Bourgeois E, Pradet A, Raymond P (1995) Molecular cloning and characterization of 6 cDNAs expressed during glucose starvation in excised maize (Zea mays L.) root tips. Plant Mol Biol 28:473–485
Choi SY, Gross KC (1994) N-Acetyl-beta-d-glucosaminidase from “Golden Delicious” apples. Phytochemistry 36:1–6
Didierjean L, Frendo P, Nasser W, Genot G, Marivet J, Burkard G (1996) Heavy-metal-responsive genes in maize: identification and comparison of their expression upon various forms of abiotic stress. Planta 199:1–8
Dyachok JV, Wiweger M, Kenne L, von Arnold S (2002) Endogenous nod-factor-like signal molecules promote early somatic embryo development in Norway spruce. Plant Physiol 128:523–533
Garcia-Casado G, Collada C, Allona I, Casado R, Pacios LF, Aragoncillo C, Gomez L (1998) Site-directed mutagenesis of active site residues in a class I endochitinase from chestnut seeds. Glycobiology 8:1021–1028
Geurts R, Franssen H (1996) Signal transduction in Rhizobium-induced nodule formation. Plant Physiol 112:447–453
Harman GE, Custis D (2006) Formulations of viable microorganisms and their method of use. US Patent WO 2007030557
Harman GE, Shoresh M (2007) The mechanisms and applications of opportunistic plant symbionts. In: Vurro M, Gressel J (eds) Novel biotechnologies for biocontrol agent enhancement and management, Springer, Amsterdam, pp 131–153
Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004a) Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2:43–56
Harman GE, Petzoldt R, Comis A, Chen J (2004b) Interactions between Trichoderma harzianum strain T22 and maize inbred line Mo17 and effects of these interactions on diseases caused by Pythium ultimum and Colletotrichum graminicola. Phytopathology 94:147–153
Hennig M, Schlesier B, Dauter Z, Pfeffer S, Betzel C, Hohne WE, Wilson KS (1992) A TIM barrel protein without enzymatic activity? Crystal structure of narbonin at 1.8-angstrom resolution. FEBS Lett 306:80–84
Hennig M, Pfeffer-Hennig S, Dauter Z, Wilson KS, Schlesier B, Nong VH (1995) Crystal structure of narbonin at 1.8-angstrom resolution. Acta Crysta D51:177–189
Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316
Huynh QK, Hironaka CM, Levine EB, Smith CE, Borgmeyer JR, Shah DM (1992) Antifungal proteins from plants. Purification, molecular cloning, and antifungal properties of chitinases from maize seed. J Biol Chem 267:6635–6640
Ishihara A, Miyagawa H, Matsukawa T, Ueno T, Mayama S, Iwamura H (1988) Induction of hydroxyanthranilate hydroxycinnamoyl transferase activity of oligo-N-acetylchitooligosaccharides in oats. Phytochemistry (Oxford) 47:929–974
Kasprzewska A (2003) Plant chitinases—regulation and function. Cell Mol Biol Lett 8:809–824
Lorito M (1998) Chitinolytic enzymes and their genes. In: Harman GE, Kubicek CP (eds) Trichoderma and Gliocladium, vol 2. Taylor and Francis, London, pp 73–99
Lorito M, Di Pietro A, Hayes CK, Woo SL, Harman GE (1993a) Antifungal, synergistic interaction between chitinolytic enzymes from Trichoderma harzianum and Enterobacter cloacae. Phytopathology 83:721–728
Lorito M, Harman GE, Hayes C, Broadway R, Tronsmo A, Woo SL, Di Pietro A (1993b) Chitinolytic enzymes produced by Trichoderma harzianum: antifungal activity of endochitinase and chitobiosidase. Phytopathology 83:302–307
Lorito M, Hayes CK, Peterbauer C, Tronsmo A, Klemsdal S, Harman GE (1993c) Antifungal chitinolytic enzymes from Trichoderma harzianum and Gliocladium virens: purification, characterization, biological activity and molecular cloning. In: Muzzarelli RAA (ed) Chitin enzymology, European Chitin Society, Ancona, pp 383–392
Lorito M, Peterbauer C, Hayes CK, Harman GE (1994) Synergistic interaction between fungal cell wall degrading enzymes and different antifungal compounds enhances inhibition of spore germination. Microbiology 140:623–629
Lorito M, Woo SL, Filippone E, Colucci G, Scala F (1996) Expression in plants of genes from mycoparasitic fungi—a new strategy for biological control of fungal diseases. International Union of Microbiological Societies (IUMS) Congresses, Jerusalem
Lorito M, Woo SL, Garcia Fernandez I, Colucci G, Harman GE, Pintor-Toro JA, Filippone E, Muccifora S, Lawrence CB, Zoina A, Tuzun S, Scala F (1998) Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proc Natl Acad Sci USA 95:7860–7865
Mauch F, Mauch-Mani B, Boller T (1988) Antifungal hydrolases in pea tissue. II. Inhibition of fungal growth by combinations of chitinase and β-1,3 glucanase. Plant Physiol 88:936–942
Nakazaki T, Tsukiyama T, Okumoto Y, Kageyama D, Naito K, Inouye K, Tanisaka T (2006) Distribution, structure, organ-specific expression and phylogenic analysis of the pathogenesis related protein-3 chitinase family in rice (Oryza sativa L.). Genome 49:619–630
Neuhaus JM (1999) Plant chitinases (PR-3, PR-4, PR-8, PR-11). In: Datta SK, Muthukrishnan S (eds) Pathogenesis-related proteins in plants, CRC press, Boca Raton, pp 77–105
Neuhaus J-M, Ahl-Goy P, Hinz U, Flores S, Meins F Jr (1991a) High-level expression of a tobacco chitinase gene in Nicotiana sylvestris. Susceptibility of transgenic plants to Cercospora nicotianae infection. Plant Mol Biol 16:141–151
Neuhaus J-M, Sticher L, Meins FJ, Boller T (1991b) A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc Natl Acad Sci 88:10362–10366
Nong VH, Schlesier B, Bassüner R, Repik A, Horstmann C, Müntz K (1995) Narbonin, a novel 2S protein from Vicia narbonensis L. seeds: cDNA, gene structure and developmentally regulated formation. Plant Mol Biol 28:61–72
Oikawa A, Itoh E, Ishihara A, Iwamura H (2003) Purification and characterization of beta-N-acetylhexosaminidase from maize seedlings. J Plant Physiol 160:991–999
Park CH, Kim S, Park JY, Ahn IP, Jwa NS, Im KH, Lee YH (2004) Molecular characterization of a pathogenesis-related protein 8 gene encoding a class III chitinase in rice. Mol Cells 17:144–150
Passarinho PA, de Vries SC (2002) Arabidopsis chitinases: a genomic survey. In: Somerville CR, Meyerowitz EM (eds) The Arabidopsis book, American Society of Plant Biologists, Rockville, pp 1–25
Patil VR, Widholm JM (1997) Possible correlation between increased vigour and chitinase activity expression in tobacco. J Exp Bot 48:1943–1950
Poulton JE, Thomas MA, Ottwell KK, McCormick SJ (1985) Partial purification and characterization of a beta-N-acetylhexosaminidase from black cherry (Prunus serotina EHRH.) seeds. Plant Sci 42:107–114
Samac DA, Shah DM (1994) Effect of chitinase antisense RNA expression on disease susceptibility of Arabidopsis plants. Plant Mol Biol 25:587–596
Schlumbaum A, Mauch F, Vogeli U, Boller T (1986) Plant chitinases are potent inhibitors of fungal growth. Nature 324:365–367
Schultze M, Staehelin C, Brunner F, Genetet I, Legrand M, Fritig B, Kondorosi E, Kondorosi A (1998) Plant chitinase/lysozyme isoforms show distinct substrate specificity and cleavage site preference towards lipochitooligosaccharide Nod signals. Plant J 16:571–580
Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, van den Elzen PJM, Cornelissen BJC (1993) Only specific tobacco (Nicotiana tabacum) chitinases and beta-1, 3-glucanases exhibit antifungal activity. Plant Physiol 101:857–863
Shoresh M, Harman GE (2008) Differential expression of maize chitinases in the presence or absence of Trichoderma harzianum strain T22. (in press)
Shoresh M, Yedidia I, Chet I (2005) Involvement of the jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology 95:76–84
Suzukawa K, Yamagami T, Ohnuma T, Hirakawa H, Kuhara S, Aso Y, Ishiguro M (2003) Mutational analysis of amino acid residues involved in catalytic activity of a family 18 chitinase from tulip bulbs. Biosci Biotechnol Biochem 67:341–346
Tiffin P (2004) Comparative evolutionary histories of chitinase genes in the genus Zea and family Poaceae. Genetics 167:1331–1340
Trudel J, Asselin A (1989) Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal Biochem 178:362–366
Trudel J, Asselin A (1994) Protein purification for microsequencing by sequential native and denaturing polyacrylamide gel electrophoresis: application to one chitinase. Anal Biochem 221:214–216
van der Holst PPG, Schlaman HRM, Spaink HP (2001) Proteins involved in the production and perception of oligosaccharides in relation to plant and animal development. Curr Opin Struct Biol 11:608–616
van Hengel AJ, Tadesse Z, Immerzeel P, Schols H, van Kammen A, de Vries SC (2001) N-Acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol 125:1880–1890
van Hengel AJ, van Kammen A, de Vries SC (2002) A relationship between seed development, Arabinogalactan-proteins (AGPs) and the AGP mediated promotion of somatic embryogenesis. Physiol Planta 114:637–644
Verburg JG, Smith CE, Lisek CA, Huynh QK (1992) Identification of an essential tyrosine residue in the catalytic site of a chitinase isolated from Zea mays that is selectively modified during inactivation with 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide. J Biol Chem 267:3886–3893
Verburg JG, Rangwala SH, Samac DA, Luckow VA, Huynh QK (1993) Examination of the role of tyrosine-174 in the catalytic mechanism of the Arabidopsis thaliana chitinase: comparison of variant chitinases generated by site-directed mutagenesis and expressed in insect cells using baculovirus vectors. Arch Biochem Biophys 300:223–230
Watanabe T, Kobori K, Miyashita K, Fujii T, Sakai H, Uchida M, Tanaka H (1993) Identification of glutamic acid 204 and aspartic acid 200 in chitinase A1 of Bacillus circulans WL-12 as essential residues for chitinase activity. J Biol Chem 268:18567–18572
Wu S, Kriz AL, Widholm JM (1994) Molecular analysis of two cDNA clones encoding acidic class I chitinase in maize. Plant Physiol 105:1097–1105
Yokoyama R, Nishitani K (2004) Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol 45:1111–1121
Acknowledgments
This research was supported in part by the US-Israel Agricultural Research and Development fund (BARD) grant US-3507-04 R and by Advanced Biological Marketing (Van Wert, OH). We thank Kristen Ondik for review and comments on the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gebhardt.
Rights and permissions
About this article
Cite this article
Shoresh, M., Harman, G.E. Genome-wide identification, expression and chromosomal location of the genes encoding chitinolytic enzymes in Zea mays . Mol Genet Genomics 280, 173–185 (2008). https://doi.org/10.1007/s00438-008-0354-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-008-0354-1