Abstract
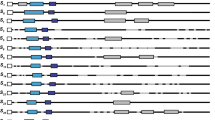
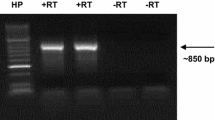
Cross-compatibility relationships in almond are controlled by a gametophytically expressed incompatibility system partly mediated by stylar RNases, of which 29 have been reported. To resolve possible synonyms and to provide data for phylogenetic analysis, 21 almond S-RNase alleles were cloned and sequenced from SP (signal peptide region) or C1 (first conserved region) to C5, except for the S 29 allele, which could be cloned only from SP to C1. Nineteen sequences (S 4 , S 6 , S 11 –S 22 , S 25 –S 29 ) were potentially new whereas S 10 and S 24 had previously been published but with different labels. The sequences for S 16 and S 17 were identical to that for S 1 , published previously; likewise, S 15 was identical to S 5 . In addition, S 4 and S 20 were identical, as were S 13 and S 19 . A revised version of the standard table of almond incompatibility genotypes is presented. Several alleles had AT or GA tandem repeats in their introns. Sequences of the 23 distinct newly cloned or already published alleles were aligned. Sliding windows analysis of Ka/Ks identified regions where positive selection may operate; in contrast to the Maloideae, most of the region from the beginning of C3 to the beginning of RC4 appeared not to be under positive selection. Phylogenetic analysis indicated four pairs of alleles had ‘bootstrap’ support > 80%: S 5 /S 10 , S 4 /S 8, S 11 /S 24 , and S 3 /S 6 . Various motifs up to 19 residues long occurred in at least two alleles, and their distributions were consistent with intragenic recombination, as were separate phylogenetic analyses of the 5′ and 3′ sections. Sequence comparison of phylogenetically related alleles indicated the significance of the region between RC4 and C5 in defining specificity.






Similar content being viewed by others
References
Beppu K, Yamane H, Yaegaki H, Yamaguchi M, Kataoka I, Tao R (2002) Diversity of S-RNase genes and S-haplotypes in Japanese plum (Prunus salicina Lindl.). J Hortic Sci Biotechnol 77:658–665
Bošković R, Tobutt KR, Batlle I, Duval H (1997) Correlation of ribonuclease zymograms and incompatibility genotypes in almond. Euphytica 97:167–176
Bošković R, Duval H, Rovira M, Tobutt KR, Romero M, Dicenta F, Batlle I (1998) Inheritance of stylar ribonucleases in two almond progenies and their correlation with self-compatibility. Acta Hort 420:118–122
Bošković R, Tobutt KR, Duval H, Batlle I, Dicenta F, Vargas FJ (1999) A stylar ribonuclease assay to detect self-compatible seedlings on almond progenies. Theor Appl Genet 99:800–810
Bošković R, Tobutt KR, Batlle I, Duval H, Martínez-Gómez P, Gradziel TM (2003) Stylar ribonucleases in almond: correlation with and prediction of incompatibility genotypes. Plant Breed 122:70–76
Channuntapipat C, Sedgley M, Collins G (2001) Sequences of the cDNAs and genomic DNAs encoding the S1, S7, S8, and Sf alleles from almond, Prunus dulcis. Theor Appl Genet 103:1115–1122
Channuntapipat C, Sedgley M, Batlle I, Arús P, Collins G (2002) Sequences of the genomic DNAs encoding the S2, S9, S10 and S23 alleles from almond, Prunus dulcis. J Hortic Sci Biotechnol 77:387–392
Channuntapipat C, Wirthensohn M, Ramesh SA, Batlle I, Arús P, Sedgley M, Collins G (2003) Identification of incompatibility genotypes in almond (Prunus dulcis Mill.) using specific primers based on the introns of the S-alleles. Plant Breed 122:164–168
Crossa-Raynaud P, Grasselly C (1985) Existence de groupes d’interstérilité chez l’amandier. Options Méditerranéennes. Ser Étud 1:43–45
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Felsenstein J (1985) Confidence limits on phylogenies: an approach using bootstrap. Evolution 39:783–791
Gagnard JM (1954) Recherches sur les caractères systématiques et les phénomènes de stérilité chez les variétés d’amandiers cultivées en Algérie. Ann Inst Agric Serv Rech Exp Agric Alger 8:1–163
Gilbert W, De Souza SJ, Long M (1997) Origin of genes. Proc Natl Acad Sci USA 94:7698–7703
Grasselly C, Olivier G (1976) Mise en évidence de quelques types autocompatibles parmi les cultivars d’amandier (P. amygdalus Batsch) de la population des Pouilles. Ann Amélior Plant 26:107–113
Horiuchi H, Yanai K, Takagi M, Yano K, Wakabayashi E, Sanda A, Mine S, Ohgi K, Irie M (1988) Primary structure of a base non-specific ribonuclease from Rhizopus niveus. J Biochem 103:408–418
Igic B, Kohn R (2001) Evolutionary relationships among self-incompatibility RNases. PNAS 98:13167–13171
Ioerger TR, Clark AG, Kao T-H (1990) Polymorphism at the self-incompatibility locus in Solanaceae predates speciation. Proc Natl Acad Sci USA 87:9732–9735
Ishimizu T, Shinkawa T, Sakiyama F, Norioka S (1998a) Primary structural features of rosaceous S-RNases associated with gametophytic self-incompatibility. Plant Mol Biol 37:931–941
Ishimizu T, Endo T, Yamaguchi-Kabata Y, Nakamura KT, Sakiyama F, Norioka S (1998b) Identification of regions in which positive selection may operate in S-RNase of Rosaceae: implication for S-allele-specific recognition sites in S-RNase. FEBS Lett 440:337–342
Kawata Y, Sakiyama F, Tamaoki H (1988) Amino-acid sequence of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem 176:683–697
Kester DE, Gradziel TM, Micke WC (1994a) Identifying pollen incompatibility groups in California almond cultivars. J Am Soc Hortic Sci 119:106–109
Kester DE, Micke WC, Viveros V (1994b) A mutation in nonpareil almond conferring unilateral incompatibility. J Am Soc Hortic Sci 119:1289–1292
López M, Mnejja M, Rovira M, Collins G, Vargas FJ, Arús P, Batlle I (2004) Self-incompatibility genotypes in almond re-evaluated by PCR, stylar ribonucleases, sequencing analysis and controlled pollinations. Theor Appl Genet 109:954–964
Ma R-C, Oliveira MM (2001) Molecular cloning of the self-incompatibility genes S1 and S3 from almond (Prunus dulcis cv. Ferragnès). Sex Plant Reprod 14:163–167
Ma R-C, Oliveira MM (2002) Evolutionary analysis of S-RNase genes from Rosaceae species. Mol Genet Genomics 267:71–78
Matsuura T, Sakai H, Unno M, Ida K, Sato M, Sakiyama F, Norioka S (2001) Crystal structure at 1.5 Å resolution of Pyrus pyrifolia pistil ribonuclease responsible for gametophytic self-incompatibility. J Biol Chem 276:45261–45269
Matton DP, Maes O, Laublin G, Xike Q, Bertrand C, Morse D, Cappadocia M (1997) Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. Plant Cell 9:1757–1766
Ortega E, Sutherland BG, Dicenta F, Bošković R, Tobutt KR (2005) Determination of incompatibility genotypes in almond using first and second intron consensus primers: detection of new S alleles and correction of reported S genotypes. Plant Breed 124:188–196
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175
Sassa H, Nishio T, Kowyama Y, Hirano H, Koba T, Ikehashi H (1996) Self-incompatibility (S) alleles of the Rosaceae encode members of a distinct class of the T2/S ribonuclease superfamily. Mol Gen Genet 250:547–557
Sonneveld T (2002) The molecular genetics of self-incompatibility in sweet cherry (Prunus avium). PhD Thesis, University of Nottingham, UK
Sonneveld T, Robbins TP, Bošković R, Tobutt KR (2001) Cloning of six cherry self-incompatibility alleles and development of allele-specific PCR detection. Theor Appl Genet 102:1046–1055
Sonneveld T, Tobutt KR, Robbins TP (2003) Allele-specific PCR detection of sweet cherry self-incompatibility (S) alleles S1 to S16 using consensus and allele-specific primers. Theor Appl Genet 107:1059–1070
Sonneveld T, Robbins TP, Tobutt KR (2006) Improved discrimination of self-incompatibility S-RNase alleles in cherry and high throughput genotyping by automated sizing of first intron polymerase chain reaction products. Plant Breed 125:305–307
Swofford DL (2003) PAUP* phylogenetic analysis using parsimony (*and other methods), Version 4. Sinauer Associates, Sunderland, MA
Sutherland BG, Robbins TP, Tobutt KR (2004) Primers amplifying a range of Prunus S-alleles. Plant Breed 123:582–584
Tamura M, Ushijima K, Sassa H, Hirano H, Tao R, Gradziel TM, Dandekar AM (2000) Identification of self-incompatibility genotypes of almond by allele-specific PCR analysis. Theor Appl Genet 101:334–349
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Ushijima K, Sassa H, Tao R, Yamane H, Dandekar AM, Gradziel TM, Hirano H (1998) Cloning and characterization of cDNAs encoding S-RNases from almond (Prunus dulcis): primary structural features and sequence diversity of the S-RNases in Rosaceae. Mol Gen Genet 260:261–268
Vieira CP, Charlesworth D, Vieira J (2003) Evidence for rare recombination at the gametophytic self-incompatibility locus. Heredity 91:262–267
Wang X, Hughes AL, Tsukamoto T, Ando T, Kao TH (2001) Evidence that intragenic recombination contributes to allelic diversity of the S-RNase gene at the self-incompatibility (S) locus in Petunia inflata. Plant Physiol 125:1012–1022
Woodward JR, Craik D, Dell A, Khoo KH, Munroe SLA, Clarke AE, Bacic A (1992) Structural analysis of the N-linked glycan chains from a stylar glycol-protein associated with expression of self-incompatibility in Nicotiana alata. Glycobiology 2:241–250
Yaegaki H, Shimada T, Moriguchi T, Hayama H, Haji T, Yamaguchi M (2001) Molecular characterization of S-RNase genes and S-genotypes in the Japanese apricot (Prunus mume Sieb. et Zucc.). Sex Plant Reprod 13:251–257
Zisovich AH, Stern RA, Sapir G, Shafir S, Goldway M (2004) The RHV region of S–RNase in the European pear (Pyrus communis) is not required for the determination of specific pollen rejection. Sex Plant Reprod 17:151–156
Acknowledgments
The authors thank F. Dicenta (CEBAS-CSIC, Spain), T.M. Gradziel (UC-Davis, USA), H. Duval (INRA-Avignon, France), and I. Batlle (IRTA-Mas Bové, Spain) for kindly providing plant material. E. Ortega acknowledges the receipt of a Postdoctoral Fellowship co-funded by the Spanish “Secretaría de Estado, de Educación y Universidades” and the European Social Fund. R. Bošković acknowledges a grant from the Mount Trust. Prunus genetic studies at East Malling Research are funded by Defra.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editorial responsibility: S. Hohmann
An erratum to this article can be found at http://dx.doi.org/10.1007/s00438-006-0168-y
Rights and permissions
About this article
Cite this article
Ortega, E., Bošković, R.I., Sargent, D.J. et al. Analysis of S-RNase alleles of almond (Prunus dulcis): characterization of new sequences, resolution of synonyms and evidence of intragenic recombination. Mol Genet Genomics 276, 413–426 (2006). https://doi.org/10.1007/s00438-006-0146-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-006-0146-4