Abstract
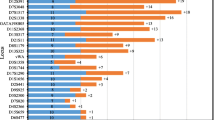
Null alleles can occur with any PCR-based STR typing system. They generally are due to deletions within the target region or primer binding sites or by primer binding site mutations that destabilize hybridization of at least one of the primers flanking the target region. Although not common, null types were detected at the DYS448 locus in seven out of 1,005 unrelated males in the Hispanic population. Of these DYS448 null types, four individuals displayed an apparent duplication at the DYS437 locus. The additional allele observed at the DYS437 locus is in actuality a smaller-sized DYS448 amplicon, which is the result of a deletion of the invariant N42 base pair domain and downstream repeats within the DYS448 locus. Thus, some DYS448 null types are not truly null. A true DYS448 null allele carried numerous primer binding site variants and a large deletion including the N42 base pair domain and surrounding or downstream repeat regions. The presence of null alleles is not a real concern for interpretation of Y STR loci evidence; current methods for interpreting Y STR profiles easily accommodate such phenomena.

Similar content being viewed by others
References
Corach D, Filgueira Risso L, Marino M, Penacino G, Sala A (2001) Routine Y-STR typing in forensic casework. Forensic Sci Int 118:131–135
Gusmão L, Butler JM, Carracedo A (2006) DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Int J Leg Med 120:191–200
Hohoff C, Dewa K, Sibbing U, Hoppe K, Forster P, Brinkmann B (2007) Y-chromosomal microsatellite mutation rates in a population sample from northwestern Germany. Int J Leg Med 121:359–363
Honda K, Roewer L, de Knijff P (1999) Male DNA typing from 25-year-old vaginal swabs using Y chromosomal STR polymorphisms in retrial request case. J Forensic Sci 44:868–872
Jobling MA, Pandya A, Tayler-Smith C (1997) The Y chromosome in forensic and paternity testing. Int J Leg Med 110:118–124
Kayser M, de Knijff P, Dieltjes P et al (1997) Applications of microsatellite-based Y-chromosome haplotyping. Electrophoresis 18:1602–1607
Krenke BE, Viculis L, Richard ML et al (2005) Validation of a male-specific, 12-locus fluorescent short tandem repeat (STR) multiplex. Forensic Sci Int 148:1–14
Mulero JJ, Chang CW, Calandro LM, Green RL, Li Y, Johnson CL, Hennessy LK (2006) Development and validation of the AmpFlSTR Yfiler PCR amplification kit: a male specific, single amplification 17 Y-STR multiplex system. J Forensic Sci 51:64–75
Park MJ, Lee HY, Chung U, Kang SC, Shin KJ (2006) Y-STR analysis of degraded DNA using reduced-size amplicons. Int J Leg Med 121:152–157
Prinz M, Boll K, Baum H, Shaler B (1997) Multiplexing of Y-chromosome specific STRs and performance of mixed samples. Forensic Sci Int 85:209–218
Sinha S, Budowle B, Chakraborty R et al (2004) Utility of the Y-STR Y-PLEXTM 6 and Y-PLEXTM 5 in forensic casework and 11 Y-STR haplotype database for three major population groups in the United States. J Forensic Sci 49:691–700
Budowle B, Sprecher CJ (2001) Concordance study on population database samples using the PowerPlexTM 16 Kit and AmpFlSTR® Profiler PlusTM Kit and AmpFlSTR® COfilerTM Kit. J Forensic Sci 46:637–641
Budowle B, Masibay A, Anderson SJ et al (2001) STR primer concordance study. Forensic Sci Int 124:47–54
Leibelt C, Budowle B, Collins P et al (2003) Identification of a D8S1179 primer binding site mutation and the validation of a primer designed to recover null alleles. Forensic Sci Int 133:220–227
Chang CW, Mulero JJ, Budowle B, Calandro LM, Hennessy LK (2006) Identification of a novel polymorphism in the X-chromosome region homologous to the DYS456 locus. J Forensic Sci 51:344–348
Collins FS, Brooks LD, Chakravarti A (1998) A DNA polymorphism discovery resource for research on human genetic variation. Genome Res 8:1229–1231
Fredman D, Munns G, Rios D et al (2004) HGVbase: a curated resource describing human DNA variation and phenotype relationships. Nucleic Acids Res 32:D516–D519
Wheeler DL, Barrett T, Benson DA et al (2005) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 33:D39–D45
Chang YM, Perumal R, Keat PY, Kuehn DL (2007) Haplotype diversity of 16 Y-chromosomal STRs in three main ethnic populations (Malays, Chinese and Indians) in Malaysia. Forens Sci Int 167:70–76
Park MJ, Kyoung-Jin S, Kim NY, Yang WI, Cho SH, Lee HY (2008) Characterization of deletions in the DYS385 flanking region and null alleles associated with AZFc microdeletions in Koreans. J Forens Sci 53:331–334
Parkin EJ, Kraayenbrink T, Opgenort JR, van Driem GL, Tuladhar NM, de Knijff P, Jobling MA (2007) Diversity of 26-locus Y-STR haplotypes in a Nepalese population sample: isolation and drift in the Himalayas. Forens Sci Int 166:176–181
Mizuno N, Nakahara H, Sekiguchi K, Yoshida K, Nakano M, Kasai K (2008) 16 Y chromosomal STR haplotypes in Japanese. Forens Sci Int 174:71–76
Roewer L, Krüger C, Willuweit S et al (2007) Y-chromosomal STR haplotypes in Kalmyk population samples. Forensic Sci Int 173:204–209
Sánchez C, Barrot C et al (2007) Haplotype frequencies of 16 Y-chromosome STR loci in the Barcelona metropolitan area population using Y-Filer kit. Forensic Sci Int 72:211–217
Redd AJ, Agellon AB, Kearney VA et al (2002) Forensic value of 14 novel STRs on the human Y chromosome. Forensic Sci Int 130:97–111
Tang JP, Hou YP, Li YB, Wu J, Zhang J, Zhang HJ (2003) Characterization of eight Y-STR loci and haplotypes in a Chinese Han population. Int J Legal Med 117:263–270
Budowle B, Giusti AM, Waye JS et al (1991) Fixed bin analysis for statistical evaluation of continuous distributions of allelic data from VNTR loci for use in forensic comparisons. Amer J Hum Genet 48:841–855
Chakraborty R, de Andrade M, Daiger SP, Budowle B (1992) Apparent heterozygote deficiencies observed in DNA typing data and their implications in forensic applications. Ann Hum Gen 56:45–57
Budowle B, Sinha SK, Lee HS, Chakraborty R (2003) Utility of Y-chromosome STR haplotypes in forensic applications. Forensic Sci Rev 15:153–164
Budowle B, Ge J, Chakraborty R (2007) Basic principles for estimating the rarity of Y-STR haplotypes derived from forensic evidence. Eighteenth International Symposium on Human Identification 2007. Promega Corporation, Madison, Wisconsin http://www.promega.com/ussymp18proc/default.htm
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Bio Evol 24:1596–1599
Acknowledgment
This is publication number 08-02 of the Laboratory Division of the Federal Bureau of Investigation. Names of commercial manufacturers are provided for identification only, and inclusion does not imply endorsement by the Federal Bureau of Investigation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Budowle, B., Aranda, X.G., Lagace, R.E. et al. Null allele sequence structure at the DYS448 locus and implications for profile interpretation. Int J Legal Med 122, 421–427 (2008). https://doi.org/10.1007/s00414-008-0258-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-008-0258-y