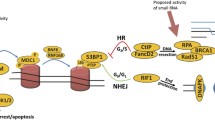
Abstract
In all living cells, DNA is constantly threatened by both endogenous and exogenous agents. In order to protect genetic information, all cells have developed a sophisticated network of proteins, which constantly monitor genomic integrity. This network, termed the DNA damage response, senses and signals the presence of DNA damage to effect numerous biological responses, including DNA repair, transient cell cycle arrests (“checkpoints”) and apoptosis. The MRN complex (MRX in yeast), composed of Mre11, Rad50 and Nbs1 (Xrs2), is a key component of the immediate early response to DNA damage, involved in a cross-talk between the repair and checkpoint machinery. Using its ability to bind DNA ends, it is ideally placed to sense and signal the presence of double strand breaks and plays an important role in DNA repair and cellular survival. Here, we summarise recent observation on MRN structure, function, regulation and emerging mechanisms by which the MRN nano-machinery protects genomic integrity. Finally, we discuss the biological significance of the unique MRN structure and summarise the emerging sequence of early events of the response to double strand breaks orchestrated by the MRN complex.






Similar content being viewed by others
References
Ajimura M, Leem SH, Ogawa H (1993) Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics 133:51–66
Akamatsu Y, Murayama Y, Yamada T, Nakazaki T, Tsutsui Y, Ohta K, Iwasaki H (2008) Molecular characterization of the role of the Schizosaccharomyces pombe nip1 + /ctp1 + gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol Cell Biol 28:3639–3651
Alani E, Padmore R, Kleckner N (1990) Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61:419–436
Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23:4868–4875
Bakkenist CJ, Kastan MB (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499–506
Barlow JH, Lisby M, Rothstein R (2008) Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell 30:73–85
Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP (2004) The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol Cell Biol 24:4151–4165
Bartek J, Bartkova J, Lukas J (2007) DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 26:7773–7779
Baudat F, de Massy B (2007) Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15:565–577
Becker E, Meyer V, Madaoui H, Guerois R (2006) Detection of a tandem BRCT in Nbs1 and Xrs2 with functional implications in the DNA damage response. Bioinformatics 22:1289–1292
Berkovich E, Monnat RJ Jr, Kastan MB (2007) Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9:683–690
Bernardi R, Pandolfi PP (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8:1006–1016
Bhaskara V, Dupre A, Lengsfeld B, Hopkins BB, Chan A, Lee JH, Zhang X, Gautier J, Zakian V, Paull TT (2007) Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol Cell 25:647–661
Boisvert FM, Dery U, Masson JY, Richard S (2005a) Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev 19:671–676
Boisvert FM, Hendzel MJ, Masson JY, Richard S (2005b) Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle 4:981–989
Borde V (2007) The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res 15:551–563
Borde V, Cobb J (2009) Double functions for the Mre11 complex during DNA double-strand break repair and replication. Int J Biochem Cell Biol 41:1249–1253
Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV (1997) A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J 11:68–76
Bressan DA, Baxter BK, Petrini JH (1999) The Mre11-Rad50-xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol 19:7681–7687
Bressan DA, Olivares HA, Nelms BE, Petrini JH (1998) Alteration of N- terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150:591–600
Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO (2008) Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135:85–96
Cahill D, Carney JP (2007) Dimerization of the Rad50 protein is independent of the conserved hook domain. Mutagenesis 22:269–274
Callebaut I, Mornon JP (1997) From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett 400:25–30
Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR 3rd, Hays L, Morgan WF, Petrini JH (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477–486
Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5:675–679
Cerosaletti KM, Desai-Mehta A, Yeo TC, Kraakman-Van Der Zwet M, Zdzienicka MZ, Concannon P (2000) Retroviral expression of the NBS1 gene in cultured Nijmegen breakage syndrome cells restores normal radiation sensitivity and nuclear focus formation. Mutagenesis 15:281–286
Chapman JR, Jackson SP (2008) Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep 9:795–801
Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE (2001) Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8:1105–1115
Chen L, Trujillo KM, Van Komen S, Roh DH, Krejci L, Lewis LK, Resnick MA, Sung P, Tomkinson AE (2005a) Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J Biol Chem 280:2620–2627
Chen PL, Liu F, Cai S, Lin X, Li A, Chen Y, Gu B, Lee EY, Lee WH (2005b) Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol 25:3535–3542
Chen L, Nievera CJ, Lee AY, Wu X (2008a) Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem 283:7713–7720
Chen YC, Chiang HY, Yang MH, Chen PM, Chang SY, Teng SC, Vanhaesebroeck B, Wu KJ (2008b) Activation of phosphoinositide 3-kinase by the NBS1 DNA repair protein through a novel activation motif. J Mol Med 86:401–412
Clerici M, Mantiero D, Lucchini G, Longhese MP (2005) The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem 280:38631–38638
Clerici M, Mantiero D, Lucchini G, Longhese MP (2006) The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep 7:212–218
Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP (2008) The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep 9:810–818
Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J (2001) Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell 8:137–147
D’Amours D, Jackson SP (2001) The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev 15:2238–2249
Daley JM, Palmbos PL, Wu D, Wilson TE (2005) Nonhomologous end joining in yeast. Annu Rev Genet 39:431–451
de Jager M, Wyman C, van Gent DC, Kanaar R (2002) DNA end-binding specificity of human Rad50/Mre11 is influenced by ATP. Nucleic Acids Res 30:4425–4431
Deng Y, Guo X, Ferguson DO, Chang S (2009) Multiple roles for MRE11 at uncapped telomeres. Nature 460:914–918
Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB (2009) Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell 34:13–25
Dery U, Coulombe Y, Rodrigue A, Stasiak A, Richard S, Masson JY (2008) A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol Cell Biol 28:3058–3069
Desai-Mehta A, Cerosaletti KM, Concannon P (2001) Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol 21:2184–2191
di Masi A, Viganotti M, Polticelli F, Ascenzi P, Tanzarella C, Antoccia A (2008) The R215W mutation in NBS1 impairs gamma-H2AX binding and affects DNA repair: molecular bases for the severe phenotype of 657del5/R215W Nijmegen breakage syndrome patients. Biochem Biophys Res Commun 369:835–840
Di Virgilio M, Gautier J (2005) Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J Cell Biol 171:765–771
Dimitrova N, de Lange T (2009) Cell cycle dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of NHEJ in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol 29(20):5552–5563
Dinkelmann M, Spehalski E, Stoneham T, Buis J, Wu Y, Sekiguchi JM, Ferguson DO (2009) Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol 16:808–813
Dong Z, Zhong Q, Chen PL (1999) The Nijmegen breakage syndrome protein is essential for Mre11 phosphorylation upon DNA damage. J Biol Chem 274:19513–19516
Dumon-Jones V, Frappart PO, Tong WM, Sajithlal G, Hulla W, Schmid G, Herceg Z, Digweed M, Wang ZQ (2003) Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res 63:7263–7269
Durocher D, Henckel J, Fersht AR, Jackson SP (1999) The FHA domain is a modular phosphopeptide recognition motif. Mol Cell 4:387–394
Dzikiewicz-Krawczyk A (2008) The importance of making ends meet: mutations in genes and altered expression of proteins of the MRN complex and cancer. Mutat Res 659:262–273
Falck J, Coates J, Jackson SP (2005) Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434:605–611
Ferreira MG, Cooper JP (2004) Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev 18:2249–2254
Frank-Vaillant M, Marcand S (2002) Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell 10:1189–1199
Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K (1998) Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J 17:6412–6425
Game JC, Mortimer RK (1974) A genetic study of x-ray sensitive mutants in yeast. Mutat Res 24:281–292
Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, Lavin MF, Gatti RA, Concannon P, Khanna K (2000) ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet 25:115–119
Ghosal G, Muniyappa K (2007) The characterization of Saccharomyces cerevisiae Mre11/Rad50/Xrs2 complex reveals that Rad50 negatively regulates Mre11 endonucleolytic but not the exonucleolytic activity. J Mol Biol 372:864–882
Gravel S, Chapman JR, Magill C, Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22:2767–2772
Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM (2006) Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev 20:34–46
Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28:739–745
Hartsuiker E, Mizuno K, Molnar M, Kohli J, Ohta K, Carr AM (2009a) Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol Cell Biol 29:1671–1681
Hartsuiker E, Neale MJ, Carr AM (2009b) Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell 33:117–123
Heikkinen K, Rapakko K, Karppinen SM, Erkko H, Knuutila S, Lundan T, Mannermaa A, Borresen-Dale AL, Borg A, Barkardottir RB et al (2006) RAD50 and NBS1 are breast cancer susceptibility genes associated with genomic instability. Carcinogenesis 27:1593–1599
Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, Bednarski JJ, Lee WL, Pandita TK, Bassing CH, Sleckman BP (2009) MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med 206:669–679
Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K et al (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180–183
Hofmann K, Bucher P (1995) The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem Sci 20:347–349
Holland IB, Blight MA (1999) ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol 293:381–399
Hopfner KP, Tainer JA (2003) Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr Opin Struct Biol 13:249–255
Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP (2000a) Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol 182:6036–6041
Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA (2000b) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101:789–800
Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA (2001) Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105:473–485
Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT et al (2002) The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418:562–566
Hopkins BB, Paull TT (2008) the p. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell 135:250–260
Horejsi Z, Falck J, Bakkenist CJ, Kastan MB, Lukas J, Bartek J (2004) Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene 23:3122–3127
Huang J, Gong Z, Ghosal G, Chen J (2009) SOSS complexes participate in the maintenance of genomic stability. Mol Cell 35:384–393
Huertas P, Jackson SP (2009) Human CtIP mediates cell-cycle control of DNA- end resection and double-strand-break repair. J Biol Chem 284(14):9558–9565
Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455:689–692
Iijima K, Muranaka C, Kobayashi J, Sakamoto S, Komatsu K, Matsuura S, Kubota N, Tauchi H (2008a) NBS1 regulates a novel apoptotic pathway through Bax activation. DNA Repair (Amst) 7:1705–1716
Iijima K, Ohara M, Seki R, Tauchi H (2008b) Dancing on damaged chromatin: functions of ATM and the RAD50/MRE11/NBS1 complex in cellular responses to DNA damage. J Radiat Res (Tokyo) 49:451–464
Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM et al (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431:1011–1017
Ivanov EL, Korolev VG, Fabre F (1992) XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics 132:651–664
Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8:37–45
Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V (2008) Mre11- Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J 27:1953–1962
Johzuka K, Ogawa H (1995) Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139:1521–1532
Kanaar R, Wyman C, Rothstein R (2008) Quality control of DNA break metabolism: in the ‘end’, it’s a good thing. EMBO J 27:581–588
Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, Kraemer KH, Shiloh Y, Bustin M (2009) Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol 11:92–96
Kobayashi J (2004) Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J Radiat Res (Tokyo) 45:473–478
Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K (2002) NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol 12:1846–1851
Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF (2006) Involvement of novel autophosphorylation sites in ATM activation. EMBO J 25:3504–3514
Krogh BO, Llorente B, Lam A, Symington LS (2005) Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics 171:1561–1570
Lee JH, Paull TT (2004) Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304:93–96
Lee JH, Paull TT (2005) ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308:551–554
Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399–409
Lee JH, Ghirlando R, Bhaskara V, Hoffmeyer MR, Gu J, Paull TT (2003) Regulation of Mre11/Rad50 by Nbs1: effects on nucleotide-dependent DNA binding and association with ataxia-telangiectasia-like disorder mutant complexes. J Biol Chem 278:45171–45181
Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28:638–651
Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA et al (2004) Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J 23:282–293
Li X, Heyer WD (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18:99–113
Li J, Lee GI, Van Doren SR, Walker JC (2000) The FHA domain mediates phosphoprotein interactions. J Cell Sci 113(Pt 23):4143–4149
Li Y, Bolderson E, Kumar R, Muniandy PA, Xue Y, Richard DJ, Seidman M, Pandita TK, Khanna KK, Wang W (2009) hSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J Biol Chem 284:23525–23531
Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB (2000) ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613–617
Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P (2007) Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28:134–146
Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699–713
Llorente B, Symington LS (2004) The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24:9682–9694
Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH (1999) Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci USA 96:7376–7381
Mahajan A, Yuan C, Lee H, Chen ES, Wu PY, Tsai MD (2008) Structure and function of the phosphothreonine-specific FHA domain. Sci Signal 1:re12
Manolis KG, Nimmo ER, Hartsuiker E, Carr AM, Jeggo PA, Allshire RC (2001) Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J 20:210–221
Mantiero D, Clerici M, Lucchini G, Longhese MP (2007) Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep 8:380–387
Maser RS, Monsen KJ, Nelms BE, Petrini JH (1997) hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol 17:6087–6096
Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y et al (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160–1166
Matsuura S, Tauchi H, Nakamura A, Kondo N, Sakamoto S, Endo S, Smeets D, Solder B, Belohradsky BH, Der Kaloustian VM et al (1998) Positional cloning of the gene for Nijmegen breakage syndrome. Nat Genet 19:179–181
Matsuzaki K, Shinohara A, Shinohara M (2008) Forkhead-associated domain of yeast Xrs2, a homolog of human Nbs1, promotes nonhomologous end joining through interaction with a ligase IV partner protein, Lif1. Genetics 179:213–225
Milne GT, Jin S, Shannon KB, Weaver DT (1996) Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol 16:4189–4198
Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455:770–774
Mirzoeva OK, Petrini JH (2001) DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol 21:281–288
Moncalian G, Lengsfeld B, Bhaskara V, Hopfner KP, Karcher A, Alden E, Tainer JA, Paull TT (2004) The rad50 signature motif: essential to ATP binding and biological function. J Mol Biol 335:937–951
Moore JK, Haber JE (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 16:2164–2173
Moreau S, Ferguson JR, Symington LS (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol 19:556–566
Moreau S, Morgan EA, Symington LS (2001) Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159:1423–1433
Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C (2005) Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437:440–443
Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J (2007) A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem 282:17330–17334
Nairz K, Klein F (1997) mre11S—a yeast mutation that blocks double-strand- break processing and permits nonhomologous synapsis in meiosis. Genes Dev 11:2272–2290
Nakada D, Matsumoto K, Sugimoto K (2003) ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev 17:1957–1962
Nakada D, Hirano Y, Sugimoto K (2004) Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol Cell Biol 24:10016–10025
Nam EA, Cortez D (2009) SOSS1/2: sensors of single-stranded DNA at a break. Mol Cell 35:258–259
Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC (2008) Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA 105:16906–16911
Niu H, Raynard S, Sung P (2009) Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev 23:1481–1486
Olson E, Nievera CJ, Lee AY, Chen L, Wu X (2007) The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem 282:22939–22952
Palmbos PL, Daley JM, Wilson TE (2005) Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol Cell Biol 25:10782–10790
Palmbos PL, Wu D, Daley JM, Wilson TE (2008) Recruitment of Saccharomyces cerevisiae Dnl4-Lif1 complex to a double-strand break requires interactions with Yku80 and the Xrs2 FHA domain. Genetics 180:1809–1819
Paull TT, Gellert M (1998) The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell 1:969–979
Paull TT, Gellert M (1999) Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev 13:1276–1288
Penkner A, Portik-Dobos Z, Tang L, Schnabel R, Novatchkova M, Jantsch V, Loidl J (2007) A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J 26:5071–5082
Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS (2009) Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 16:819–824
Raymond WE, Kleckner N (1993) RAD50 protein of S.cerevisiae exhibits ATP- dependent DNA binding. Nucleic Acids Res 21:3851–3856
Richard DJ, Bolderson E, Cubeddu L, Wadsworth RI, Savage K, Sharma GG, Nicolette ML, Tsvetanov S, McIlwraith MJ, Pandita RK et al (2008) Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 453:677–681
Rodriguez MC, Songyang Z (2008) BRCT domains: phosphopeptide binding and signaling modules. Front Biosci 13:5905–5915
Rupnik A, Grenon M, Lowndes N (2008) The MRN complex. Curr Biol 18:R455–R457
Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450:509–514
Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K et al (2007) Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell 130:863–877
Shima H, Suzuki M, Shinohara M (2005) Isolation and characterization of novel xrs2 mutations in Saccharomyces cerevisiae. Genetics 170:71–85
Shimada M, Sagae R, Kobayashi J, Habu T, Komatsu K (2009) Inactivation of the Nijmegen breakage syndrome gene leads to excess centrosome duplication via the ATR/BRCA1 pathway. Cancer Res 69:1768–1775
Shiotani B, Zou L (2009) Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell 33:547–558
Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice. Cell Res 18:134–147
Sinha KM, Unciuleac MC, Glickman MS, Shuman S (2009) AdnAB: a new DSB-resecting motor-nuclease from mycobacteria. Genes Dev 23:1423–1437
Slijepcevic P (2006) The role of DNA damage response proteins at telomeres—an “integrative” model. DNA Repair (Amst) 5:1299–1306
Soutoglou E, Misteli T (2008) Activation of the cellular DNA damage response in the absence of DNA lesions. Science 320:1507–1510
Squatrito M, Gorrini C, Amati B (2006) Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol 16:433–442
Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM (1999) The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99:577–587
Stracker TH, Theunissen JW, Morales M, Petrini JH (2004) The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 3:845–854
Sun Y, Xu Y, Roy K, Price BD (2007) DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol 27:8502–8509
Takeda S, Nakamura K, Taniguchi Y, Paull TT (2007) Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell 28:351–352
Tauchi H, Kobayashi J, Morishima K, Matsuura S, Nakamura A, Shiraishi T, Ito E, Masnada D, Delia D, Komatsu K (2001) The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50·hMRE11·NBS1 complex DNA repair activity. J Biol Chem 276:12–15
Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, vanHeems D, Ito E, Nakamura A, Sonoda E et al (2002) Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 420:93–98
Taylor AM, Groom A, Byrd PJ (2004) Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair (Amst) 3:1219–1225
The International Nijmegen Breakage Syndrome Study Group (2000) Nijmegen Breakage Syndrome. The International Nijmegen Breakage Syndrome Study Group. Arch Dis Child 82:400–406
Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno KI, Ohta K, Uritani M, Ushimaru T et al (2003) Competition between the Rad50 complex and the Ku Heterodimer reveals a role for exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23:5186–5197
Tommiska J, Seal S, Renwick A, Barfoot R, Baskcomb L, Jayatilake H, Bartkova J, Tallila J, Kaare M, Tamminen A et al (2006) Evaluation of RAD50 in familial breast cancer predisposition. Int J Cancer 118:2911–2916
Trujillo KM, Sung P (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem 276:35458–35464
Trujillo KM, Yuan SS, Lee EY, Sung P (1998) Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem 273:21447–21450
Tsubouchi H, Ogawa H (2000) Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell 11:2221–2233
Tsukamoto Y, Mitsuoka C, Terasawa M, Ogawa H, Ogawa T (2005) Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol Biol Cell 16:597–608
Uanschou C, Siwiec T, Pedrosa-Harand A, Kerzendorfer C, Sanchez-Moran E, Novatchkova M, Akimcheva S, Woglar A, Klein F, Schlogelhofer P (2007) A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J 26:5061–5070
Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T (1998) Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705–716
Usui T, Ogawa H, Petrini JH (2001) A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell 7:1255–1266
Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y (2003) Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22:5612–5621
van der Linden E, Sanchez H, Kinoshita E, Kanaar R, Wyman C (2009) RAD50 and NBS1 form a stable complex functional in DNA binding and tethering. Nucleic Acids Res 37(5):1580–1588
Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ et al (1998) Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93:467–476
Wang Z, Cummins JM, Shen D, Cahill DP, Jallepalli PV, Wang TL, Parsons DW, Traverso G, Awad M, Silliman N et al (2004) Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res 64:2998–3001
Weterings E, Chen DJ (2008) The endless tale of non-homologous end-joining. Cell Res 18:114–124
Williams RS, Williams JS, Tainer JA (2007) Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 85:509–520
Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G et al (2008) Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135:97–109
Wiltzius JJ, Hohl M, Fleming JC, Petrini JH (2005) The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol 12:403–407
Wu G, Lee WH (2006) CtIP, a multivalent adaptor connecting transcriptional regulation, checkpoint control and tumor suppression. Cell Cycle 5:1592–1596
Wu X, Ranganathan V, Weisman DS, Heine WF, Ciccone DN, O’Neill TB, Crick KE, Pierce KA, Lane WS, Rathbun G et al (2000) ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405:477–482
Wu L, Luo K, Lou Z, Chen J (2008) MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci USA 105:11200–11205
Xiao Y, Weaver DT (1997) Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res 25:2985–2991
Xie A, Kwok A, Scully R (2009) Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 16:814–818
Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C et al (1999) Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J 18:6619–6629
Yang YG, Saidi A, Frappart PO, Min W, Barrucand C, Dumon-Jones V, Michelon J, Herceg Z, Wang ZQ (2006) Conditional deletion of Nbs1 in murine cells reveals its role in branching repair pathways of DNA double-strand breaks. EMBO J 25:5527–5538
Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG (2009) The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol Biol Cell 20:2351–2360
You Z, Chahwan C, Bailis J, Hunter T, Russell P (2005) ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 25:5363–5379
Yu X, Baer R (2000) Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J Biol Chem 275:18541–18549
Yu X, Chen J (2004) DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol 24:9478–9486
Yu X, Chini CC, He M, Mer G, Chen J (2003) The BRCT domain is a phospho-protein binding domain. Science 302:639–642
Yu X, Fu S, Lai M, Baer R, Chen J (2006) BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev 20:1721–1726
Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E (2007) SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell 27:149–162
Yun MH, Hiom K (2009) CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459:460–463
Zha S, Boboila C, Alt FW (2009) Mre11: roles in DNA repair beyond homologous recombination. Nat Struct Mol Biol 16:798–800
Zhang Y, Zhou J, Lim CU (2006) The role of NBS1 in DNA double strand break repair, telomere stability, and cell cycle checkpoint control. Cell Res 16:45–54
Zhu J, Petersen S, Tessarollo L, Nussenzweig A (2001) Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol 11:105–109
Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134:981–994
Acknowledgements
The authors thank Sunichi Takeda for his authorisation to cite unpublished results, Serge Gravel and Karl Peter Hopfner for critical reading of the manuscript and John Eykelenboom for his comments. The work in Noel Lowndes laboratory is supported by Cancer Research Ireland under grant CR105GRE, European Commission Integrated project “DNA Repair” contract number 512113 and Science Foundation Ireland Principal Investigator award 07/IN1/B958.
Author information
Authors and Affiliations
Corresponding authors
Additional information
During the preparation of these proofs, two articles have been published describing the NBS1 crystal structure of both FHA domain and BRCT repeats of human and S. pombe cells. The results presented in these publications suggest an important role in phospheptide binding for both of these domains. Scott Williams et al. Cell, 139 (1) 87–99, Nbs1 Flexibly Tethers Ctp1 and Mre11-Rad50 to Coordinate DNA Double-Strand Break Processing and Repair Lloyd et al. Cell, 139 (1) 100–111, A Supramodular FHA/BRCT-Repeat Architecture Mediates Nbs1 Adaptor Function in Response to DNA Damage.
Rights and permissions
About this article
Cite this article
Rupnik, A., Lowndes, N.F. & Grenon, M. MRN and the race to the break. Chromosoma 119, 115–135 (2010). https://doi.org/10.1007/s00412-009-0242-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-009-0242-4