Abstract
There are currently about 80 different DNA tests available for mutations that are associated with inherited disease in the domestic dog, and as the tools available with which to dissect the canine genome become increasingly sophisticated, this number can be expected to rise dramatically over the next few years. With unrelenting media pressure focused firmly on the health of the purebred domestic dog, veterinarians and dog breeders are turning increasingly to DNA tests to ensure the health of their dogs. It is ultimately the responsibility of the scientists who identify disease-associated genetic variants to make sensible choices about which discoveries are appropriate to develop into commercially available DNA tests for the lay dog breeder, who needs to balance the need to improve the genetic health of their breed with the need to maintain genetic diversity. This review discusses some of the factors that should be considered along the route from mutation discovery to DNA test and some representative examples of DNA tests currently available.
Similar content being viewed by others
Introduction
In December 2004 the $30 million project funded by the National Human Genome Research Institute (NHGRI) in the United States to sequence the entire dog genome was completed and the results made publicly available. The NHGRI made the decision to fund such an expensive undertaking because it recognised the dog as an unrivalled model organism with which to study the genetics of a wide range of inherited traits, including inherited disease, behaviour, and morphology. Although the NHGRI’s motives for sequencing all the DNA in the dog were primarily human-centric, the findings that have emerged from the canine genome project, and that will continue to materialise for many years to come, have profound implications for both veterinary and human medical research. Most importantly, the pace at which genetic variants associated with inherited canine diseases have been identified has increased dramatically and will continue to do so as the tools available to dissect the genetic basis of canine inherited traits become increasingly more sophisticated.
Studies over the last decade, aimed at understanding the genetic basis of inherited traits in the domestic dog, have revealed mutations in dozens of genes, some of them novel, which are now worthy candidates for further study in other species, including man. In addition to mutations associated with Mendelian traits, an ever-increasing number of loci that are associated with genetically complex conditions are being reported. Many of these will almost certainly shed light on the development of similar conditions in other species as well.
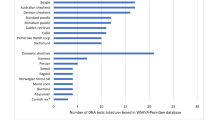
A better understanding of the nature and function of some of the genes identified in the dog that are associated with equivalent conditions in man will no doubt lead, in the long term, to improved diagnosis and treatment for human patients. It is also possible that many of these downstream benefits will eventually also be applicable to dogs. But by far the most likely and immediate improvement to the health of domestic dogs will derive from the DNA tests that are routinely made available once a disease-associated mutation has been identified. Currently, DNA tests are available for over 80 different canine mutations, a number that can be expected to increase very quickly in the coming years, and over 120 breeds are able to take advantage of at least one DNA test. A list of DNA tests available for dogs at the time of writing is given in Table 1, although it should be noted that new DNA tests are becoming available very rapidly so readers should check with individual testing laboratories for a complete list of tests available. Alternative resources for finding lists of currently available DNA tests are http://www.thekennelclub.org.uk/item/2108, http://www.offa.org/dna_alltest.html, and http://www.akcchf.org/canine-health/genetic-tests/. This article does not attempt to discuss every test available. Rather it discusses some of the considerations that should be taken into account to successfully translate scientific findings into robust and useful tools for the lay dog breeder to use, and in so doing it uses a few representative examples of DNA tests that are currently available.
The domestic dog, inherited disease, and DNA testing
Intense selection, high levels of inbreeding, the extensive use of a limited number of sires, and genetic isolation are all hallmarks of modern breeds of domestic dog. It is widely agreed that part of the collateral damage from these practices is that purebred dogs have a greater risk of suffering from genetically simple inherited disorders than their cross-breed cousins. Intense media interest continues to apply pressure on both dog breeders and the veterinary profession to improve the health of purebred dogs, the result being that these stakeholders are turning increasingly to DNA tests to assist with both breeding decisions and also the diagnostic process. Some dog breeders are exceptionally well informed when it comes to genetics and possess a very good understanding of the inherited disorders that affect their breed(s). But many are not, and they rely on their general practice veterinarian for information, who also may not have a good knowledge of the majority of inherited conditions that affect all the different breeds they encounter.
It is, therefore, ultimately the responsibility of the scientists who identify genetic variants associated with canine inherited disorders to exercise prudence when they make DNA tests available. Purebred dog populations, by definition, are genetically compromised, with many having an unequivocal need to maximise diversity. Breeders are increasingly using DNA tests to shape their breeding decisions, with the need to reduce the frequency of deleterious mutations being balanced with the need to maintain genetic diversity. There is a critical need, therefore, for full and transparent information, written in lay terms, explaining (1) the mode of inheritance and penetrance of specific mutations, (2) the risk of disease associated with specific genotypes, (3) the frequency of each mutation within different breed populations, and (4) the breeds that are genuinely at risk, so that dogs are not unnecessarily eliminated from the breeding population. The existence of phenocopies within the breed, if known, should also be documented.
DNA testing of companion animals is unregulated, and so one of the few ways in which potential “customers” can judge the merit of any DNA test is if a description of the mutation on which the test is based has been accepted by a peer-reviewed scientific publication. However, the data that establish a genetic variant as a provocative candidate for downstream, comparative studies (and therefore worthy of publication) is not always the same information that a dog owner needs to discern whether they should use the DNA test and what decisions to make in light of the results. It is therefore not sufficient for a DNA test provider merely to provide a hyperlink to a peer-reviewed publication without backing it up with some layman-friendly explanation.
It is worth noting that once a mutation has been reported in the scientific literature there is little stopping private, for-profit organisations from offering a genetic test based on a published finding, irrespective of the opinion of the scientists who made the discovery. Therefore, suggesting that researchers have absolute control over which DNA tests are offered might be an oversimplification in some cases. Nevertheless, scientists should certainly be encouraged to offer “user-friendly” advice on how to use genetic tests and to work with dog breed clubs and organisations to disseminate this advice. Perhaps they should also be willing to suggest when consideration of a particular disease-associated variant to shape breeding decisions is inappropriate.
DNA tests for recessive mutations
The vast majority of DNA tests currently available are for autosomal recessive mutations associated with Mendelian or single-gene traits. Some of these traits are morphological, such as coat colour, but most are for disease mutations, many of which are severe and debilitating. Providing advice to breeders for these tests is generally very straightforward; essentially all dogs can be safely bred, regardless of their genotype, provided both the sire and the dam have been tested and carriers and genetically affected dogs are mated only to dogs that are clear of the mutation. Often, the initial reaction of breeders is to avoid breeding with all but homozygous wild-type (+/+) dogs, and indeed many members of the veterinary community who were indoctrinated to never breed affected dogs condone this view. However, if a mutation is frequent within a breed, breeders should be counselled to include carriers in the breeding population, for at least a generation, to avoid reducing diversity unnecessarily. Recent data from the 1,000 genomes project revealed that humans carry, on average, between 250 and 300 recessive mutations and at least 50 mutations previously associated with inherited disorders, and it seems reasonable to assume the average dog will carry the same burden of disease-associated variants (Durbin et al. 2010). Expecting breeding dogs to be clear of all risk alleles, therefore, is unrealistic and will severely jeopardise breed diversity.
Recessive mutations for late-onset conditions are notoriously difficult to eliminate if a DNA test is not available, and, in the absence of effective selective pressure, can become common within a breed. Nevertheless, robust data regarding mutation frequency are often unavailable, possibly because the data are rarely required for publication, and as the number of DNA tests available increases, it will become increasingly important for veterinarians and breeders to be able to sensibly prioritise tests for specific breeds. DNA test providers should be prepared to provide accurate information regarding the frequency of a specific mutation in relevant breeds, both when a DNA test is initially made available and at regular intervals, so progress toward the genetic improvement of the breed can be monitored. Obtaining such estimates might require sampling of additional individual dogs from a cross section of the gene pool because DNA samples collected during a genetic investigation are usually biased toward affected individuals and closely related dogs.
Numerous DNA tests are available for autosomal recessive mutations, far too many to describe individually, so this review summarises two representative examples, along with background behind their identification. Both mutations have been reported in the scientific literature and both are relevant to a large number of breeds. The mutations are those for progressive rod cone degeneration (prcd) (Zangerl et al. 2006) and primary lens luxation (PLL) (Farias et al. 2010).
Progressive rod cone degeneration (prcd) is a late-onset form of progressive retinal atrophy (PRA) that affects multiple breeds. Prior to characterization of this disease at the molecular level, elegant interbreed crosses were undertaken to determine that the phenotypically similar diseases that were segregating in multiple breeds, including the miniature poodle, the English and American cocker spaniels, the Labrador retriever, the Australian cattle dog, the Nova Scotia duck tolling retriever, and the Portuguese water dog, were in fact allelic (Aguirre and Acland 1988, 2006). However, when prcd-affected dogs were mated to PRA-affected dogs of the Border Collie, Basenji, and Italian greyhound breeds, the progeny were normal, indicating that these breeds are affected by genetically distinct forms of disease. The prcd locus was mapped to a large region on CFA9 in 1998 (Acland et al. 1998), before the canine genome sequence was available and while tools with which to investigate the canine genome were relatively unsophisticated compared to those available today. The whole-genome radiation panels that were available at the time, and that would have been useful to investigate any other region of the genome, did not significantly help to locate the mutation because they were both TK1 selected (Priat et al. 1998), and since TK1 was tightly linked to the prcd locus, it was difficult to order positional candidate genes within the prcd critical region. However, the fact that a genetically identical disease segregated in so many breeds proved to be invaluable as it allowed the use of linkage equilibrium mapping across affected breeds to considerably narrow the prcd-associated region (Goldstein et al. 2006) and led to the eventual identification of a single nucleotide substitution in the second codon of a previously unknown gene that is now known to be the cause of prcd in over 30 different breeds (Zangerl et al. 2006). A DNA test for the prcd mutation is provided by OptiGen (www.optigen.com), a service company cofounded by ophthalmologists Gregory Acland and Gustavo Aguirre to provide DNA-based diagnoses and information about inherited diseases of dogs. The company provides breeding advice as well as comprehensive, breed-specific information for owners regarding, for example, the average age of onset of prcd in specific breeds and whether genetically distinct forms of PRA are known to exist in the same breed.
The mutation for PLL is another example of a mutation that segregates in multiple breeds, all of which can take advantage of a DNA test. PLL is an inherited deficiency of the lens suspensory apparatus, the zonule, which is a system of fibres that suspend the lens from the ciliary body, maintaining it on the visual axis and in contact with the anterior surface of the vitreous body. In dogs affected with PLL, ultrastructural abnormalities of the zonular fibres are already evident at 20 months of age (Curtis 1983), long before the lens luxation that typically occurs when the dogs are 3–8 years old, as a result of degeneration and breakdown of the zonules that cause the lens to be displaced from its normal position within the eye (Curtis 1990; Curtis and Barnett 1980; Curtis et al. 1983; Morris and Dubielzig 2005). In the majority of cases the dislocated lens will pass into the anterior chamber where its presence is likely to cause acute glaucoma. The condition has been recognized as a canine familial disorder for more than 100 years (Gray 1909, 1932) and is encountered at high frequency in several terrier breeds and in some other breeds with probable terrier coancestry (Curtis 1990; Curtis and Barnett 1980; Curtis et al. 1983; Morris and Dubielzig 2005; Willis et al. 1979). In 2010 a mutation in ADAMTS17 was described as the cause of PLL in three breeds, the Miniature Bull terrier, the Lancashire Heeler, and the Jack Russell terrier. The mutation is a G→A substitution at c.1473+1, which destroys a splice donor recognition site in intron 10 and causes exon skipping that results in a frameshift and the introduction of a premature termination codon (Farias et al. 2010). A subsequent publication, targeted at a veterinarian audience, reports 14 additional breeds in which the identical mutation segregates, and documents the frequency of the mutation in a subset of breeds (Gould et al. 2011). The great majority of PLL-affected dogs are homozygous for the mutation, but a small minority are heterozygous, leading to speculation that carriers, of some breeds at least, might be at increased risk of developing the condition compared to dogs that are homozygous for the wild-type allele (Farias et al. 2010). A DNA test for the ADAMTS17 substitution is available via each of the two research groups that collaborated to identify the mutation, the Animal Health Trust (www.aht.org.uk) and the University of Missouri, via The Orthopaedic Foundation for Animals (http://www.offa.org/dnatesting/pll.html), as well as a number of other DNA-testing providers that have used the published data to develop their own tests. Both the Animal Health Trust and The Orthopaedic Foundation for Animals provide extensive information regarding the clinical aspects of PLL, the risk to dogs with each of the three possible genotypes of developing PLL, and also breeding advice for owners. The mutation is frequent in several breeds, so advice is provided that counsels breeders to include heterozygotes in the breeding population to avoid pushing breeds through a genetic bottleneck. Both prcd and PLL are examples of where rigorous scientific investigations have provided the data for DNA tests that are provided in a suitable context for dog owners and breeders to use to full advantage.
Validating which breeds a specific DNA test should be usefully offered to is an important consideration for the DNA test provider, and many DNA test providers, including the Animal Health Trust and OptiGen (N. Holmes and S. Pearce-Kelling, personal communications), have a policy of restricting all tests to only those breeds in which the mutation has previously been identified and is also associated with disease. The risk associated with specific mutations might vary, depending on the genetic background, so simply establishing that a mutation is segregating within a breed does not necessarily justify making the test available to that breed.
Both prcd and PLL are genetically “simple” conditions, that is, the mutations are completely penetrant and homozygous dogs invariably develop the associated condition during their lives. Examples are emerging of recessive conditions for which the associated mutation is not completely penetrant, indicating that other factors, either genetic or environmental, play a role in the development of the disease. Researchers need to provide dog breeders with specific counselling for such conditions to ensure that they appreciate that not all dogs that are homozygous for disease-associated variants will develop clinical signs during their lives, but that they will pass these mutations onto their offspring (who may inherit different modifier alleles and be affected).
One example of a disease that segregated as a Mendelian trait within an inbred research colony but appeared genetically more complex in the general pet population is a form of retinal degeneration described in the miniature long-haired dachshund (MLHD). The disease was originally described as an early-onset, autosomal recessive PRA, with all affected dogs within an inbred research colony displaying ophthalmologic abnormalities that were detectable by electroretinogram (ERG) by 6 weeks of age and by fundoscopy at 25 weeks of age. The dogs were invariably blind by the time they were 2 years of age (Curtis and Barnett 1993). A subsequent ERG study identified an initial reduction of the cone photoreceptor function which led to the condition being reclassified as a cone–rod dystrophy (CRD) rather than a rod-led PRA, and the disease was termed cord1 for cone–rod degeneration 1 (Turney et al. 2007). The same condition has also been referred to by others as crd4 for cone–rod degeneration 4 (Aguirre and Acland 2006). Using the same colony of dogs, cord1 was mapped to a large region on CFA15, and a mutation in RPGRIP1 was identified that cosegregated completely with cord1 in the research colony (Mellersh et al. 2006a). The mutation is a 44-bp insertion of an A29 tract flanked by a 15-bp duplication in exon 2 of the gene, which creates a frameshift and introduces a premature stop codon early in exon 3. Mutations in RPGRIP1 have been associated with Leber congenital amaurosis (LCA) (Dryja et al. 2001), retinitis pigmentosa (RP) (Booij et al. 2005), and CRD (Hameed et al. 2003) in humans, as well as inherited retinal abnormalities in mice (Zhao et al. 2003), which suggests that it plays an important role in visual function. Within the research colony of MLHDs there was complete correlation between the RPGRIP1 genotype and phenotype of the dogs with respect to their cord1 phenotype, whereas in the pet MLHD population this was not the case (Miyadera et al. 2009). Outside of the colony there was considerable variation in the age of onset of retinal degeneration in dogs that were homozygous for the RPGRIP1 insertion (RPGRIP1 −/−), which has also been identified in other breeds, including the English Springer Spaniel (ESS) and the Beagle. However, all RPGRIP1 −/− Beagles and MLHDs showed reduced or absent ERG cone responses, even in the absence of ophthalmoscopic abnormalities, a finding that has also been corroborated by Busse et al. (2011). Together, these findings suggest that additional mutations which modify the age of onset of ophthalmoscopic abnormalities associated with the RPGRIP1 mutation are involved. Because the original research colony used was developed from a very small number of dogs, it is likely that the colony was fixed for these additional loci which, therefore, went undetected until the more outbred pet population was investigated. A recent association study using RPGRIP1 −/− MLHDs that had either early- or late-onset cord1 has indeed revealed a second locus that segregates with early-onset disease (K. Miyadera, Cambridge, 2010, personal communication), indicating that early-onset CRD in MLHDs is more likely to be a digenic condition and that the RPGRIP1 insertion alone causes a late-onset CRD, although ERG abnormalities may be detected early in life. The RPGRIP1 insertion is very frequent within both MLHD and ESS populations, probably due to the lack of effective selection against the late-onset CRD with which it is associated. Breeders should thus be mindful to breed with carriers so that the genetic diversity of their breed(s) is not unduly compromised and to consider the RPGRIP1 genotype of their dogs alongside all the other factors they use to weigh a dog’s breeding potential.
Two additional examples of recessive mutations that are not completely penetrant but are highly associated with disease (and therefore sound candidates for a DNA test) are those for degenerative myelopathy (DM) in the Boxer, Cardigan Welsh Corgi, Chesapeake Bay Retriever, German Shepherd, Kerry Blue Terrier, Pembroke Welsh Corgi, Rhodesian Ridgeback, and Standard Poodle and hyperuricosuria.
DM is a severe, incurable disease of the spinal cord where demyelination and axonal loss contribute to degeneration of the white matter of the spinal cord resulting in progressive paralysis. The disease has an insidious onset, typically between 8 and 14 years of age, so affected dogs may well have been bred well before their clinical signs develop. The mutation that is tested for, a single-nucleotide missense mutation in SOD1, greatly increases an individual dog’s risk of developing DM, although a minority of dogs that carry two copies of the mutation remain free of clinical signs associated with the condition during their lifetime. It is suspected that there are additional mutations and/or environmental factors that modify the effects of the DM mutation and explain why some dogs remain healthy (Awano et al. 2009). The orthopaedic foundation for animals (OFA) makes it very clear on their DNA-testing website that the DM carrier status of dogs should be considered alongside other factors, such as breed type and temperament, and that breeders should not “over-emphasize DNA test results” (http://www.offa.org/dnatesting/dmbreederguide.html).
Another example of a disease-associated risk factor that is incompletely penetrant and for which there is a commercially available DNA test is the mutation associated with hyperuricosuria, or elevated levels of uric acid in the urine. This trait predisposes dogs to form stones in their bladders, or sometimes kidneys, which often have to be removed surgically and can be difficult to treat. The associated variant, a missense mutation in SLC2A9, is associated with hyperuricosuria in several different breeds, including the Dalmatian (a breed that is fixed for the mutation), but as with DM, not all dogs that are homozygous for the mutation develop urate stones, indicating that other factors are involved (Bannasch et al. 2008; Karmi et al. 2010). The DNA tests for these and other disease-associated variants provide dog breeders with invaluable tools with which to reduce the frequency of inherited diseases from breeds at risk, and, provided the mutation being tested for is highly associated with the disease in question, it is difficult to justify not making such DNA tests available to the public. Provided full information is available to enable the nonscientist to understand the level of risk involved, that the mutation is not fully penetrant and that other factors might contribute to the development of disease, then breeds will certainly benefit in the long term from the availability of such tests.
DNA tests for dominant mutations
Dominant mutations obviously present dog breeders with a different dilemma from recessive mutations; because all offspring that inherit a disease-associated dominant mutation will develop clinical signs at some stage during their lives, it is harder to justify breeding with animals that carry dominant mutations. Depending on the frequency of the mutation within a population, tests for dominant mutations have the potential to result in more dogs being prevented from breeding, and consequently do more damage to the genetic diversity of a breed, than DNA tests for recessive mutations. As a result, researchers have an even greater responsibility to limit the availability of DNA tests for dominant mutations to those that are highly associated with disease. They need to exercise sensible restraint for mutations that have incomplete penetrance (particularly if the level of penetrance is very low), especially if the disease-associated mutation is frequent within a breed. There are several DNA tests currently available that are based on dominant or codominant mutations, including the RHO mutation for an autosomal dominant form of PRA in the English Mastiff offered by OptiGen (www.optigen.com) (Kijas et al. 2002), the mutation in HSF4 that is associated with hereditary cataract in the Australian Shepherd that is offered by the Animal Health Trust (www.AHT.org.uk) (Mellersh et al. 2006b, 2009), and the polymorphisms associated with renal dysplasia in multiple breeds that is offered by DoGenes (www.dogenes.com) (Whiteley et al. 2011).
Complex conditions
Genetically complex conditions that result from mutations in multiple genes or the interaction between genes and the environment represent a vastly different scenario from genetically simple traits. Individual polymorphisms will invariably be identified that increase an individual dog’s risk of developing an associated condition but cannot predict with absolute certainty whether the dog will become clinically affected. Since the canine genome sequence became available, four different, increasingly dense, whole-genome SNP arrays have been developed [the 27 K(v1) and 50 K(v2) canine Affymetrix SNP chips and the Illumina CanineSNP20 and CanineHD BeadChips], providing increasingly sensitive means with which to identify regions of the genome associated with complex traits of interest. Several reports have been published that identify regions of the canine genome associated with complex traits (Bannasch et al. 2010; Wilbe et al. 2010), and precise variants underlying these associations will almost certainly be identified in the near future. Associations between specific genes and complex conditions are obviously important and will make profound contributions toward our understanding of gene function and disease aetiology, but they present the scientist with a dilemma when it comes to deciding whether to make a DNA test available based on risk factors. There is a need for dog owners and breeders to balance the desire to breed for “genetic health” by eliminating disease-associated mutations from their breed with the need to maintain genetic diversity. As stated already, most dog breeders are not generally in a position to objectively evaluate the specific level of risk associated with a given mutation from a scientific publication. There will be a tendency for all mutations described as “risk factors” to be considered similarly, regardless of the level of risk they convey, and once a DNA test becomes commercially available, many breeders will have their dogs tested and avoid breeding with dogs that carry that mutation. It is, therefore, irresponsible of DNA test providers to offer tests for isolated mutations that are only weakly associated with disease; these mutations are likely to be minor modifiers of more major risk factors and eliminating them may reduce genetic diversity without dramatically reducing the prevalence of disease. Increasingly, research to identify canine disease-associated mutations is being funded by dog-oriented organisations, and researchers need to resist pressure to make DNA tests available prematurely to compensate these stakeholders for their financial and moral support.
Summary
Genetic tools available with which to dissect the canine genome are becoming increasingly sophisticated, and the number of disease-associated genetic variants that will be identified over the next few years will undoubtedly increase dramatically. These mutations will form the basis of extremely valuable tools that dog owners and breeders will clamour to use to eliminate both Mendelian and complex inherited conditions from their beloved breeds. If the public is to maintain confidence in these new tools, scientists must be responsible custodians of their findings, and make DNA tests commercially available only if they can be accompanied by straightforward, transparent, user-friendly advice on how they should be used to reduce the frequency of disease without reducing genetic diversity unnecessarily.
References
Acland GM, Ray K, Mellersh CS, Gu W, Langston AA, Rine J, Ostrander EA, Aguirre GD (1998) Linkage analysis and comparative mapping of canine progressive rod–cone degeneration (prcd) establishes potential locus homology with retinitis pigmentosa (RP17) in humans. Proc Natl Acad Sci USA 96:3048–3053
Aguirre GD, Acland GM (1988) Variation in retinal degeneration phenotype inherited at the prcd locus. Exp Eye Res 46:663–687
Aguirre GD, Acland GM (2006) Models, mutants and man: searching for unique phenotypes and genes in the dog model of inherited retinal degeneration. In: Ostrander EA, Giger U, Lindblad-Toh K (eds) The dog and its genome. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 291–325
Awano T, Johnson GS, Wade CM, Katz ML, Johnson GC, Taylor JF, Perloski M, Biagi T, Baranowska I, Long S, March PA, Olby NJ, Shelton GD, Khan S, O’Brien DP, Lindblad-Toh K, Coates JR (2009) Genome-wide association analysis reveals a SOD1 mutation in canine degenerative myelopathy that resembles amyotrophic lateral sclerosis. Proc Natl Acad Sci USA 106:2794–2799
Bannasch D, Safra N, Young A, Karmi N, Schaible RS, Ling GV (2008) Mutations in the SLC2A9 gene cause hyperuricosuria and hyperuricemia in the dog. PLoS Genet 4:e1000246
Bannasch D, Young A, Myers J, Truve K, Dickinson P, Gregg J, Davis R, Bongcam-Rudloff E, Webster MT, Lindblad-Toh K, Pedersen N (2010) Localization of canine brachycephaly using an across breed mapping approach. PLoS ONE 5:e9632
Booij JC, Florijn RJ, ten Brink JB, Loves W, Meire F, van Schooneveld MJ, de Jong PT, Bergen AA (2005) Identification of mutations in the AIPL1, CRB1, GUCY2D, RPE65, and RPGRIP1 genes in patients with juvenile retinitis pigmentosa. J Med Genet 42:e67
Busse C, Barnett KC, Mellersh CS, Adams VJ (2011) Ophthalmic and cone derived electrodiagnostic findings in outbred miniature long-haired dachshunds homozygous for a RPGRIP1 mutation. Vet Ophthalmol 14:146–152
Curtis R (1983) Aetiopathological aspects of inherited lens dislocation in the Tibetan Terrier. J Comp Pathol 93:151–163
Curtis R (1990) Lens luxation in the dog and cat. Vet Clin North Am Small Anim Pract 20:755–773
Curtis R, Barnett KC (1980) Primary lens luxation in the dog. J Small Anim Pract 21:657–668
Curtis R, Barnett KC (1993) Progressive retinal atrophy in miniature longhaired dachshund dogs. Br Vet J 149:71–85
Curtis R, Barnett KC, Lewis SJ (1983) Clinical and pathological observations concerning the aetiology of primary lens luxation in the dog. Vet Rec 112:238–246
Dryja TP, Adams SM, Grimsby JL, McGee TL, Hong DH, Li T, Andreasson S, Berson EL (2001) Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am J Hum Genet 68:1295–1298
Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA (2010) A map of human genome variation from population-scale sequencing. Nature 467:1061–1073
Farias FH, Johnson GS, Taylor JF, Giuliano E, Katz ML, Sanders DN, Schnabel RD, McKay SD, Khan S, Gharahkhani P, O’Leary CA, Pettitt L, Forman OP, Boursnell M, McLaughlin B, Ahonen S, Lohi H, Hernandez-Merino E, Gould DJ, Sargan D, Mellersh CS (2010) An ADAMTS17 splice donor site mutation in dogs with primary lens luxation. Invest. Ophthalmol Vis Sci 51:4716–4721
Goldstein O, Zangerl B, Pearce-Kelling S, Sidjanin DJ, Kijas JW, Felix J, Acland GM, Aguirre GD (2006) Linkage disequilibrium mapping in domestic dog breeds narrows the progressive rod–cone degeneration interval and identifies ancestral disease-transmitting chromosome. Genomics 88:541–550
Gould D, Pettitt L, McLaughlin B, Holmes N, Forman O, Thomas A, Ahonen S, Lohi H, O’Leary C, Sargan D, Mellersh C (2011) ADAMTS17 mutation associated with primary lens luxation is widespread among breeds. Vet Ophthalmol 14:1–7
Gray H (1909) The diseases of the eye in domesticated animals. Vet Rec 21:678
Gray H (1932) Some medical and surgical conditions in the dog and cat. Vet Rec 12:1–10
Hameed A, Abid A, Aziz A, Ismail M, Mehdi SQ, Khaliq S (2003) Evidence of RPGRIP1 gene mutations associated with recessive cone–rod dystrophy. J Med Genet 40:616–619
Karmi N, Brown EA, Hughes SS, McLaughlin B, Mellersh CS, Biourge V, Bannasch DL (2010) Estimated frequency of the canine hyperuricosuria mutation in different dog breeds. J Vet Intern Med 24:1337–1342
Kijas JW, Cideciyan AV, Aleman TS, Pianta MJ, Pearce-Kelling SE, Miller BJ, Jacobson SG, Aguirre GD, Acland GM (2002) Naturally occurring rhodopsin mutation in the dog causes retinal dysfunction and degeneration mimicking human dominant retinitis pigmentosa. Proc Natl Acad Sci USA 99:6328–6333
Mellersh CS, Boursnell ME, Pettitt L, Ryder EJ, Holmes NG, Grafham D, Forman OP, Sampson J, Barnett KC, Blanton S, Binns MM, Vaudin M (2006a) Canine RPGRIP1 mutation establishes cone–rod dystrophy in miniature longhaired dachshunds as a homologue of human Leber congenital amaurosis. Genomics 88:293–301
Mellersh CS, Pettitt L, Forman OP, Vaudin M, Barnett KC (2006b) Identification of mutations in HSF4 in dogs of three different breeds with hereditary cataracts. Vet Ophthalmol 9:369–378
Mellersh CS, McLaughlin B, Ahonen S, Pettitt L, Lohi H, Barnett KC (2009) Mutation in HSF4 is associated with hereditary cataract in the Australian Shepherd. Vet Ophthalmol 12:372–378
Miyadera K, Kato K, Aguirre-Hernandez J, Tokuriki T, Morimoto K, Busse C, Barnett K, Holmes N, Ogawa H, Sasaki N, Mellersh CS, Sargan DR (2009) Phenotypic variation and genotype-phenotype discordance in canine cone–rod dystrophy with an RPGRIP1 mutation. Mol Vis 15:2287–2305
Morris RA, Dubielzig RR (2005) Light-microscopy evaluation of zonular fiber morphology in dogs with glaucoma: secondary to lens displacement. Vet Ophthalmol 8:81–84
Priat C, Hitte C, Vignaux F, Renier C, Jiang Z, Jouquand S, Cheron A, Andre C, Galibert F (1998) A whole-genome radiation hybrid map of the dog genome. Genomics 54:361–378
Turney C, Chong NH, Alexander RA, Hogg CR, Fleming L, Flack D, Barnett KC, Bird AC, Holder GE, Luthert PJ (2007) Pathological and electrophysiological features of a canine cone–rod dystrophy in the miniature longhaired dachshund. Invest Ophthalmol Vis Sci 48:4240–4249
Whiteley MH, Bell JS, Rothman DA (2011) Novel allelic variants in the canine cyclooxgenase-2 (Cox-2) promoter are associated with renal dysplasia in dogs. PLoS ONE 6:e16684
Wilbe M, Jokinen P, Truve K, Seppala EH, Karlsson EK, Biagi T, Hughes A, Bannasch D, Andersson G, Hansson-Hamlin H, Lohi H, Lindblad-Toh K (2010) Genome-wide association mapping identifies multiple loci for a canine SLE-related disease complex. Nat Genet 42:250–254
Willis MB, Curtis R, Barnett KC, Tempest WM (1979) Genetic aspects of lens luxation in the Tibetan terrier. Vet Rec 104:409–412
Zangerl B, Goldstein O, Philp AR, Lindauer SJ, Pearce-Kelling SE, Mullins RF, Graphodatsky AS, Ripoll D, Felix JS, Stone EM, Acland GM, Aguirre GD (2006) Identical mutation in a novel retinal gene causes progressive rod–cone degeneration in dogs and retinitis pigmentosa in humans. Genomics 88:551–563
Zhao Y, Hong DH, Pawlyk B, Yue G, Adamian M, Grynberg M, Godzik A, Li T (2003) The retinitis pigmentosa GTPase regulator (RPGR)-interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc Natl Acad Sci USA 100:3965–3970
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Laboratories
AHT | www.aht.org.uk (UK) |
Alfort School of Veterinary Medicine | www.labradorcnm.com (France) |
Antagene | www.antagene.com (France) |
Auburn University | www.vetmed.auburn.edu (USA) |
Cornell University | www.web.vet.cornell.edu/ (USA) |
Finnzymes | www.finnzymes.fi (Finland) |
Genindexe | www.genindexe.com (France) |
Genetic Technologies Ltd | www.geneticscienceservices.com (Australia) |
GenMARK | www.genmarkag.com (USA) |
Genoscoper | |
HealthGene | www.healthgene.com (Canada) |
Laboklin | www.laboklin.co.uk (UK) |
OFA | www.offa.org (USA) |
OptiGen | www.optigen.com (USA) |
PennGen | www.vet.upenn.edu (USA) |
University of California | |
University of Missouri | |
University of Utrecht | email: P.A.J.Leegwater@vet.uu.nl (Holland) |
Van Haeringen | email: info@vhlgenetics.com (Holland) |
VDA | www.vetdnacenter.com (USA) |
VetGen | www.vetgen.com (USA) |
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Mellersh, C. DNA testing and domestic dogs. Mamm Genome 23, 109–123 (2012). https://doi.org/10.1007/s00335-011-9365-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-011-9365-z