Abstract
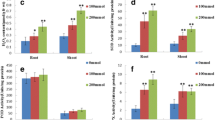
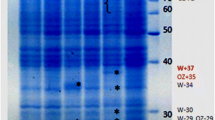
Soluble proteins from the salt-tolerant Rhizobium etli strain EBRI 26 were separated by two-dimensional (2D) gel electrophoresis and visualised by Commassie staining. Six proteins are highly expressed after induction by 4% NaCl compared to the non-salt-stressed cells. These proteins have pI between 5 and 5.5 and masses of approximately 22, 25, 40, 65, 70, and 95 kDa. These proteins were analysed by Matrix-assisted laser adsorption ionization time of flight (MALDI-TOF) after digestion with trypsin. Despite having very good peptide mass fingerprint data, these proteins could not be identified, because the genome sequence of R. etli is not yet published. In a second approach, soluble proteins from salt-induced or non-salt-induced cultures from R. etli strain EBRI 26 were separately labelled with different fluorescent cyano-dyes prior to 2D difference in gel electrophoresis. Results revealed that 49 proteins are differentially expressed after the addition of sodium chloride. Fourteen proteins are overexpressed and 35 were downregulated. The genome of Sinorhizobium meliloti, a closely related species to R. etli, has been published. Similar experiments using Sinorhizobium meliloti strain 2011 identified four overexpressed and six downregulated proteins. Among the overexpressed protein is a carboxynospermidin decarboxylase, which plays an important role in the biosynthesis of spermidin (polyamine). The enzyme catalase is among the downregulated proteins. These proteins may play a role in salt tolerance.



Similar content being viewed by others
Literature Cited
Asch F, Dingkuhn M, Wittstock C, Doerffling K (1999) Sodium and potassium uptake of rice panicles as affected by salinity and season in relation to yield components. Plant Soil 207:133–145
Aziz A, Martin-Tanguy J, Larher F (1999) Salt stress-induced proline accumulation and changes in tyramine and ployamine levels are linked to ionic adjustment in tomato leaf discs. Plant Sci 145:83–91
Bolaños L, El-Hamdaoui A, Bonilla I (2003) Recovery of development and functionality of nodules and plant growth in salt-stressed Pisum sativum-Rhizobium leguminosarum symbiosis by boron and calcium. J Plant Physiol 160:1493–1497
Botsford JL (1990) Analysis of protein expression in response to osmotic stress in Escherichia coli. FEMS Microbiol Lett 72:355–360
Bradford M (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cavalcanti FR, Oliveira JTA, Martins-Miranda AS, Viégas RA, Silveira JAG (2004) Superoxide dismutase, catalase and peroxidase activities do not confer protection against oxidative damage in salt-stressed cowpea leaves. New Phytologist 163:563–571
Delgado MJ, Garrido JM, Ligero F, Lluch C (1993) Nitrogen fixation and carbon metabolism by nodules and bacteroids of pea plants under sodium chloride stress. Physiol Plantarum 89:824–829
Die Lajudie P, Laurent-Fulele E, Willems A, Tork U, Coopman R, Collins MD, Kersters K, Dreyfus B, Gillis M (1998) Allorhizobium undicola gen. nov., sp. nov., nitrogen fixing bacteria that efficiently nodulate Neptunia natans in Senegal. Int J Syst Bacteriol 48:1277–1290
Djordjevic MA (2004) Sinorhizobium meliloti metabolism in the root nodule: A proteomic perspective. Proteomics 4:1859–1872
Drolet G, Dumbroff EB, Legge RL, Thompson JE (1986) Radical scavenging properties of polyamines. Phytochemistry 25:367–371
El-Sheikh EAE, Wood M (1995) Nodulation and N2 fixation by soybean inoculated with salt-tolerant rhizobia or salt-sensitive bradyrhizobia in saline soil. Soil Biol Biochem 27:657–661
Encarnación S, Guzmán Y, Dunn MF, Hernández M, Vargas Maria C, Mora J (2003) Proteom analysis of aerobic and fermentative metabolism in Rhizobium etli CE3. Proteomics 3:1077–1085
Gade D, Thiermann J, Markowsky D, Rabus R (2003) Evaluation of two-dimensional difference gel electrophoresis for protein profiling. J Mol Microbiol Biotech 5:240–251
Georgiev GI, Atkias CA (1993) Effects of salinity on N2 fixation, nitrogen metabolism and export and diffusive conductance of cowpea root nodules. Symbiosis 15:239–255
Galibert F, Finan TM, Long SR, Pühler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, Bothe G, Boutry M, Bowser L, Buhrmester J, Cadieu E, Capela D, Chain P, Cowie A, Davis RW, Dréano S, Federspiel NA, Fisher RF, Gloux S, Godrie T, Goffeau A, Golding B, Gouzy J, Gurjal M, Hernandez-Lucas I, Hong A, Huizar L, Hyman RW, Jones T, Kahn D, Kahn ML, Kalman S, Keating DH, Kiss E, Komp C, Lelaure V, Masuy D, Palm C, Peck MC, Pohl TM, Portetelle D, Purnelle B, Ramsperger U, Surzycki R, Thébault P, Vandenbol M, Vorhälter FJ, Weidner S, Wells DH, Wong K, Yeh KC, Batut J (2001) The composite genome of the legume symbiont Sinorhizosbium meliloti. Science 293:668–672
Galibert F, Barloy-Hubler F, Capela D, Gouzy J (2000) Sequencing the Sinorhizobium meliloti genome. DNA Sequence 11:207–210
Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W (2000) The current state of two dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21:1037–1053
Hellman V, Wernstedt C, Gonez J, Heldin CH (1995) Improvement of an (In-Gel) digestion procedure for the micro preparation of internal protein fragments for amino acid sequencing. Anal Biochem 224:421–455
Hu Y, Schmidhalter U (1998) Spatial distributions of inorganic ions and sugars contributing to osmotic adjustment in the elongating wheat leaf under saline soil conditions. Austr J Plant Physiol 25:591–597
Kasinathan V, Wingler A (2004) Effect of reduced arginine decarboxylase activity on salt tolerance and on polyamine formation during salt stress in Arabidopsis thaliana. Physiol Plantarum 121:101–107
Lauber WM, Carroll JA, Dufield DR, Kiesel JR, Radabaugh MR, Malone JP (2001) Mass spectrometry compatibility of two-dimensional gel protein stains. Electrophoresis 22:906–918
Liu K, Fu H, Bei Q, Luan S (2000) Inward potassium channel in guard cells as a target for polyamine regulation of stomatal movements. Plant Physiol 124:1315–1325
Martinez-Romero E, Caballero-Mellado J (1996) Rhizobium phylogenies and bacterial genetic. Crit Rev Plant Sci 15:113–140
Nogales J, Campos R, BenAbdelkhalek H, Olivares J, Lluch C, Sanjuan J (2002) Rhizobium tropici genes involved in free-living salt tolerance are required for the establishment of efficient nitrogen-fixing symbiosis with Phaseolus vulgaris. Mol Plant-Microbe Interact 15:225–232
Oren A (1999) Bioenergetic aspects of Halophilism. Microbiol Mol Biol Rev 63:334–348
Patton WF (2002) Detection technologies in proteom analysis. J Chromatogr B 771:3–31
Peick B, Graumann P, Schmid R, Marahiel M, Werner D (1999) Differential pH-induced proteins in Rhizobium tropici CIAT 899 and Rhizobium etli CIAT 611. Soil Biol Biochem 31:189–194
Rabus R, Brüchert V, Amann J, Könneke M (2002) Physiological response to temperature changes of the marine, sulphate-reducing bacterium Desulfobacterium autotrophicum. FEMS Microbiol Ecol 42:409–417
Rai R, Nasar SKT, Singh SJ, Prasad V (1985) Interaction between Rhizobium strains and lentil (Lens culinaris) genotypes under salt-stress. J Agric Sci 104:199–205
Rüberg S, Tian ZX, Krol E, Linke B, Meyer F, Wang Y, Pühler A, Weidner S, Becker A (2003) Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression. J Biotech 106:255–268
Sairam RK, Tyagi A (2004) Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci 86:407–421
Shamseldin A, Werner D (2005) High salt and high pH tolerance of new isolated R. etli strains from Egyptian soils. Curr Microbiol 50:11–16
Smith LT, Smith GM (1989) An osmoregulated dipeptide in stressed Rhizobium meiloti. J Bacteriol 171:4714–4717
Soussi M, Ocaña A, Lluch C (1998) Effects of salt stress on growth, photosynthesis and nitrogen fixation in chick-pea (Cicer arietinum L.). J Exp Botany 49:1329–1337
TeChien C, Maundu J, Cavaness J, Dandurand LM, Orser CS (1992) Characterization of salt-tolerant and salt-sensitive mutants of Rhizobium leguminosarum biovar viciae strain C1204b. FEMS Microbiol Lett 90:135–140
Tejera NA, Campos R, Sanjuan J, Lluch C (2004) Nitrogenase and antioxidant enzyme activities in Phaseolus vulgaris nodules formed by Rhizobium tropici isogenic strains with varying tolerance to salt stress. J Plant Physiol 161:329–338
Tiburcio AF, Besford RT, Capell T, Borrel A, Testillano PS, Risueño MC (1994) Mechanisms of polyamine action during senescence responses induced by osmotic stress. J Exp Botany 45:1789–1800
Unni S, Rao KK (2001) Protein and lipopolysaccharide profiles of a salt-sensitive Rhizobium sp. and its exopolysaccharide-deficient mutant. Soil Biol Biochem 33:111–115
Werner D (1999) Körnerleguminosen: Biologische Stickstoff-Fixierung. In: Keller ER, Hanus H, Heyland K-U (Hrgs) Handbuch des Pflanzenbaues, Band 3 Knollen- und Wurzelfrüchte, Körner- und Futterleguminosen. Stuttgart, Verlag Eugen Ulmer, S. 554–562
Werner D, Wilcockson J, Zimmerman E (1975) Adsorption and selection of rhizobia by ion exchange papers. Arch Microbiol 105:27–32
Zahran HH, Zaghloul TI, Ahmad MS, Ahmad ES (2004) A salt-tolerant and nitrogen fixing mutant of Rhizobium leguminosarum bv. viciae isolated from Egypt. Nitrogen Fixation Conferrence, Toulouse, France, 24–27 July, Abstract p 132
Zahran HH (1999) Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev 63:968–989
Zahran HH, Rasanen LA, Karsisto M, Lindström K (1994) Alteration of lipopolysaccharide and protein profiles in SDS-PAGE of rhizobia by osmotic and heat stress. World J Microbiol Biotechnol 10:100–105
Acknowledgments
We would like to thank Prof. Dr. Klaus Lingelbach for permitting use of his protein analysis facility. We also thank Jürgen Thiermann from Amersham Biosciences for his assistance with the DIGE technology and analysis software, World Lab Organization for a short grant, and the EU for support through the INCO-DEV project ICA4-CT-2001-10057.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shamseldin, A., Nyalwidhe, J. & Werner, D. A Proteomic Approach Towards the Analysis of Salt Tolerance in Rhizobium etli and Sinorhizobium meliloti Strains. Curr Microbiol 52, 333–339 (2006). https://doi.org/10.1007/s00284-005-6472-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-005-6472-7