Abstract
Rhamnolipids are glycolipidic biosurfactants produced by various bacterial species. They were initially found as exoproducts of the opportunistic pathogen Pseudomonas aeruginosa and described as a mixture of four congeners: α-L-rhamnopyranosyl-α-L-rhamnopyranosyl-β-hydroxydecanoyl-β-hydroxydecanoate (Rha-Rha-C10-C10), α-L-rhamnopyranosyl-α-L-rhamnopyranosyl-β-hydroxydecanoate (Rha-Rha-C10), as well as their mono-rhamnolipid congeners Rha-C10-C10 and Rha-C10. The development of more sensitive analytical techniques has lead to the further discovery of a wide diversity of rhamnolipid congeners and homologues (about 60) that are produced at different concentrations by various Pseudomonas species and by bacteria belonging to other families, classes, or even phyla. For example, various Burkholderia species have been shown to produce rhamnolipids that have longer alkyl chains than those produced by P. aeruginosa. In P. aeruginosa, three genes, carried on two distinct operons, code for the enzymes responsible for the final steps of rhamnolipid synthesis: one operon carries the rhlAB genes and the other rhlC. Genes highly similar to rhlA, rhlB, and rhlC have also been found in various Burkholderia species but grouped within one putative operon, and they have been shown to be required for rhamnolipid production as well. The exact physiological function of these secondary metabolites is still unclear. Most identified activities are derived from the surface activity, wetting ability, detergency, and other amphipathic-related properties of these molecules. Indeed, rhamnolipids promote the uptake and biodegradation of poorly soluble substrates, act as immune modulators and virulence factors, have antimicrobial activities, and are involved in surface motility and in bacterial biofilm development.
Similar content being viewed by others
Introduction
Surfactants are chemical compounds that (as entailed by their name) display surface activity. They have predilection for interfaces of dissimilar polarities (liquid–air or liquid–liquid) and are soluble in both organic (non-polar) and aqueous (polar) solvents. This property originates from their amphiphilic (or amphipathic) structures, which comprise both hydrophilic (head) and hydrophobic groups (tail) (Desai and Banat 1997). Biosurfactants are surfactants of biological origin. Microorganisms like bacteria, yeasts, and fungi are known to produce various types of biosurfactants. Their hydrophilic part is usually composed of sugars, amino acids, or polar functional groups like carboxylic acid groups. The hydrophobic part is typically an aliphatic hydrocarbon chain of β-hydroxy fatty acids (Lang and Wullbrandt 1999). Surfactants are generally classified according to the charge carried by their polar groups (head) into cationic, anionic, amphoteric, and nonionic.
Biosurfactants are attracting much attention because they represent ecological alternatives to their synthetic counterparts: they exhibit lower toxicity, potentially high activities, and stability at extremes of temperature, pH, and salinity. They have a wide variety of structures, and they can be produced from renewable feedstocks by a wide variety of microorganisms. Most importantly, they are biodegradable, making them environmentally friendly, “green” chemicals.
Biosurfactants are classified according to their molecular structure into mainly glycolipids (e.g., rhamnolipids (RLs) and sophorolipids), lipopeptides (e.g., surfactin), polymeric biosurfactants (e.g., emulsan and alasan), fatty acids ((e.g., 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs)), and phospholipids (e.g., phosphatidylethanolamine) (Desai and Banat 1997; Lang and Wullbrandt 1999).
Rhamnolipids are surface-active glycolipids. They have been intensively investigated and extensively reviewed (Maier and Soberón-Chávez 2000; Nitschke et al. 2005; Ochsner et al. 1996; Soberón-Chávez 2004; Soberón-Chávez et al. 2005). However, two questions about this class of biosurfactants are emerging as important topics that have yet been poorly reviewed. First, what is the intrinsic, natural role of RLs for the producing organisms? Second, what actually are these producers? Indeed, while the opportunistic pathogen Pseudomonas aeruginosa has traditionally been considered the primary RL-producing microorganism, many other bacterial species, especially in recent years, have been reported to produce RLs as well. Still, apart from some clear cases, it appears that the ability to produce RLs is in fact restricted to a limited number of species, and that many reports are anecdotal. Although initially only four RL species had been described, the development of more sensitive analytical techniques has revealed the co-production of a far wider variety of RL congeners. Thus, this review focuses on the diversity of reported RL bacterial producers, the chemical structure of identified RLs, and the various physiological functions and roles attributed to RLs for the producing bacteria.
Diversity of chemical structures of rhamnolipids
The initial discovery of RLs goes back to 1946 when Bergström et al. (1946a, b) reported an oily glycolipid produced by Pseudomonas pyocyanea (now P. aeruginosa) after growth on glucose that was named pyolipic acid and whose structural units were identified as l-rhamnose and β-hydroxydecanoic acid (Hauser and Karnovsky 1954; Jarvis and Johnson 1949). Jarvis and Johnson (1949) further elucidated the structure of a RL isolated from P. aeruginosa and showed that it was composed of two β-hydroxydecanoic acids linked through a glycosidic bond to two rhamnose moieties, with the two β-hydroxy fatty acid portions linked through an ester bond while the disaccharide portion contained a putative 1,3-glycosidic linkage. Edwards and Hayashi (1965) rather found that the linkage between the two rhamnose moieties is an α-1,2-glycosidic linkage, as determined by periodate oxidation and methylation. Based on that, they chemically described this RL as 2-O-α-1,2-L-rhamnopyranosyl-α-L-rhamnopyranosyl-β-hydroxydecanoyl-β-hydroxydecanoate (di-RL, structure 39 in Table 1). This was the first discovered glycolipid containing a link between a sugar and a hydroxylated fatty acid residue (Shaw 1970).
Later on, Itoh et al. (1971) isolated and identified a new RL congener produced concurrently with the aforementioned di-RL by several strains of P. aeruginosa after growth on n-paraffin, and it was identified as α-L-rhamnopyranosyl-β-hydroxydecanoyl-β-hydroxydecanoate (mono-RL, structure 13 in Table 1). Moreover, they postulated that this mono-RL is the precursor of the di-RL (Itoh et al. 1971), although this had been suggested previously (Burger et al. 1963). A few years later, two additional RL congeners were identified in cultures of Pseudomonas sp. DSM 2874 grown as resting cells on n-alkanes or glycerol (Syldatk et al. 1985). These were similar to the previously identified mono- and di-RLs but containing only one β-hydroxydecanoic acid side chain (mono-lipidic RLs; structures 3 and 31 in Table 1). These mono-lipidic RLs were also detected in trace concentrations in continuous P. aeruginosa cultures (Gruber et al. 1993). Nevertheless, it is unlikely that these mono-lipidic congeners require special culture conditions to be produced since many recent studies have identified the four RL congeners produced by various P. aeruginosa in batch-mode production. For instance, Arino et al. (1996) have reported the following composition in a P. aeruginosa batch culture: 67% di-rhamno-di-lipid, 22% mono-rhamno-di-lipid, 9% di-rhamno-mono-lipid, and less than 3% of mono-rhamno-mono-lipid.
Furthermore, two unusual di-lipidic RLs have been identified with a further acylation with a 2-decenoyl group at the free 2-hydroxyl group of the rhamnose moieties, in cultures of a hydrocarbon-utilizing Pseudomonas sp. after growth on n-paraffin (structures 29 and 58 in Table 1; Yamaguchi et al. 1976). These two congeners have been also described by Lang and Wagner (1987). Since then, however, very few teams have reported these two congeners, a possible indication that they are strain-specific (Ishigami et al. 1987a, b, 1996; Yamaguchi et al. 1978). Finally, two novel RLs were described as methyl esters of the free carboxylic groups of two previously identified di-lipidic RLs (structures 28 and 57 in Table 1; Hirayama and Kato 1982). These two methylated congeners are the only known nonionic RLs.
Apart from these eight RL congeners identified until the mid-1980s, the last decade of the twentieth century has witnessed the discovery of a large number of new RLs that paralleled the implementation of more sensitive analytical techniques, such as high-performance liquid chromatography (HPLC), mass spectrometry (MS), and HPLC/MS. Rendell et al. (1990) identified five new di-RL congeners from cultures of a clinical isolate of P. aeruginosa. These RLs had aliphatic chains of variable lengths and some contained an unsaturation (C8-C10, C10-C8, C10-C12, C12-C10, and C10-C12:1 as illustrated in structures 36, 37, 44, 45, and 42, respectively, in Table 1). They also reported corresponding mono-RL congeners (structures 10, 11, 18, 19, and 16 in Table 1) using HPLC/MS. A rhamnolipid congener in which the sum of the carbons in the side chains amounted to C24 was identified in addition to the previous homologues by Zhang and Miller (1994) in a P. aeruginosa isolate; however, the exact length of each β-hydroxy-fatty acid was not mentioned. Déziel et al. (1999, 2000) reported, in addition to the previously identified congeners, the di-RL congeners containing C8-C8, C8-C12:1, C12:1-C8, C12:1-C10, C12:1-C12, C10-C14:1, C12-C12, C8, C12:1, and C12 moieties (structures 35, 40, 41, 43, 48, 46, 49, 30, 32, and 33, respectively, in Table 1), as well as other new mono-RL congeners containing C8-C8, C12:1-C10, C8, and C12 moieties (structures 7, 17, 2, and 5, respectively, in Table 1) produced by a P. aeruginosa strain grown on mannitol or naphthalene as carbon sources. All in all, they reported a total number of 28 RL congeners, many of which were present in only minute concentrations.
New congeners are still regularly reported. Abalos et al. (2001) have identified new mono-RL congeners with the polyunsaturated β-hydroxy-fatty acid chains C8:2 and C12:2 (structures 1 and 4 in Table 1) in cultures of a P. aeruginosa isolate, yet their concentrations were low. Interestingly, these congeners were produced in significant amounts by a different P. aeruginosa isolate (AT10) in another study (Haba et al. 2003a). Additional mono-RL homologues, containing C10-C14:1, C12-C12:1, and C10-C14 (structures 20, 21, and 22 in Table 1) were detected in cultures of Pseudomonas chlororaphis strain NRRL B-30761 (Gunther et al. 2005). Recently, other di-RL congeners with mono-unsaturation as C10-C10:1 and C12-C12:1 (structures 38 and 47 in Table 1) were also identified (Sharma et al. 2007). Moreover, new mono-RLs containing C8-C10:1, C10-C10:1, and C12-C12 moieties (structures 8, 12, and 23 in Table 1) were identified in culture of the P. aeruginosa isolate LBI, although the first two congeners only represented less than 1% of all the RLs detected (Nitschke et al. 2009). Recently, other mono-RL congeners containing C14:2 and C10:1-C8 moieties (structures 6 and 9 in Table 1) have been identified in cultures of a hyper-producing P. aeruginosa mutant (MIG-N146) (Guo et al. 2009).
All the previously mentioned RL congeners and homologues were identified in various isolates of Pseudomonas species, essentially P. aeruginosa. However, other RL homologues produced by non-Pseudomonas bacterial isolates have also been reported. For example, Pajarron et al. (1993) have identified new di-RL congeners produced by a Pseudomonas glumae isolate, a species later renamed as Burkholderia glumae (Urakami et al. 1994). The major di-RL congeners they identified contained a C14-C14 moiety; other minor congeners containing C12-C14, C14-C12, C14-C16, and C16-C14 moieties were also identified (structures 52, 50, 51, 53, and 54 in Table 1). The major di-RL congener containing C14-C14 was also identified in other species of Burkholderia, such as the pathogen Burkholderia pseudomallei, but as the sole RL congener (Häussler et al. 1998; Howe et al. 2006). However, Andrä et al. (2006) identified in addition to the Rha-Rha-C14-C14 congener two other RL congeners with mono- and tri-acylation with β-hydroxy-tetradecanoic acid: Rha-Rha-C14 and Rha-Rha-C14-C14-C14 (stated as unpublished data), respectively (structures 34 and 56 in Table 1) produced by Burkholderia plantarii. Apart from the work of Pajarron et al. (1993), the few above-mentioned reports on RLs produced by Burkholderia sp. have identified only the Rha-Rha-C14-C14 congener. However, Dubeau et al. (2009) have recently identified new mono- and di-RL homologues from Burkholderia thailandensis grown on glycerol or vegetable oil, with the following combinations of fatty acids: C10-C12, C12-C12, C12-C14, C14-C14, C14-C16, and C16-C16, among which, the major congeners are still those with C14-C14 side chains. Other congeners were also found in trace amonts (Dubeau et al. 2009). Thus, the newly identified RL congeners to be added to the pool of known RLs are the mono-RLs with C12-C14, C14-C14, C14-C16, and C16-C16 (structures 24, 25, 26, and 27 in Table 1) as well as the di-RL with C16-C16 (structures 55 in Table 1). Finally, they also identified a range of di-RL congeners produced by B. pseudomallei carrying the same combinations of C12, C14, and C16 fatty acids as seen with B. thailandensis (Dubeau et al. 2009).
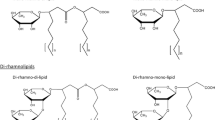
Thus, in view of the structures mentioned above, RLs can be described as follows: they are glycosides composed of rhamnose moieties (glycon part) and lipid moieties (aglycon part) linked to each other via a O-glycosidic linkage. The glycon part is composed of one (mono-RLs) or two (di-RLs) rhamnose moieties linked to each other through a α-1,2-glycosidic linkage. The aglycon part, however, is mainly one or two (in few cases three) β-hydroxy fatty acid chains (saturated, mono-, or poly-unsaturated and of chain length varying from C8 to C16) linked to each other through an ester bond formed between the β-hydroxyl group of the distal (relative to the glycosidic bond) chain with the carboxyl group of the proximal chain (Fig. 1). In most cases, the carboxyl group of the distal β-hydroxy fatty acid chain remains free; few congeners, however, have this group esterified with a short alkyl group. Similarly, the 2-hydroxyl group of the distal (relative to the glycosidic bond) rhamnose group remains mostly free, although in some rare homologues it can be acylated with long chain alkenoic acid.
Chemical structure of the first identified rhamnolipid; simply named as α-L-rhamnopyranosyl-α-L-rhamnopyranosyl-β-hydroxydecanoyl-β-hydroxydecanoate and symbolized as Rha-Rha-C10-C10. Its IUPAC names: (R)-3-{(R)-3-[2-O-(α-L-rhamnopyranosyl)-α-L-rhamnopyranosyl]oxydecanoyl}oxydecanoate or the synonym name: (R)-3-((R)-3-((2R,3R,4R,5R,6S)-4,5-dihydroxy-6-methyl-3-((2S,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yloxy)tetrahydro-2H-pyran-2-yloxy)decanoyloxy)decanoic acid
Variations in the chemical structures of bacterially produced RLs give rise to a large pool of RL homologues that approaches 60 structures (Table 1). The differences among these homologues come from modifications in the glycon and/or the aglycon parts, taking in consideration that variation in the aglycon part contribute largely to the biodiversity of RLs. Although few general rules apply, Déziel et al. (1999, 2000) previously noted that for RL isomers containing two fatty acid moieties of different chain lengths, the predominant (at least twice more abundant) isomeric congeners will be those with the shorter chain adjacent to the sugar. This proportion is even larger when one of the two fatty acid moieties contains a double bond, i.e., the most prevalent isomeric congener will be that with the shorter saturated fatty acid chain adjacent to the sugar while its isomeric congener with the unsaturated longer chain adjacent to the sugar is found at much lower concentrations (Déziel et al. 2000) or, in other studies, is completely absent (Sharma et al. 2007). Concerning the β-hydroxyl groups of the fatty acid chains, they were reported to be strictly present in the R-configuration (Bauer et al. 2006; Schenk et al. 1997).
Diversity of biological producers
Pseudomonas species have long been the main sources of RLs, with P. aeruginosa being considered the primary producing species. However, as noted above, many isolates from other bacterial species of varying distance in their taxonomical classification are increasingly reported to be also RL producers. First, many Pseudomonas species other than P. aeruginosa have been reported to produce RLs (Gunther et al. 2005, 2006; Onbasli and Aslim 2009). Some belong to the same taxonomical class of Pseudomonas (Gammaproteobacteria) and order (Pseudomonadales) but are not part of the Pseudomonadacae family. For instance, two Acinetobacter calcoaceticus isolates (order: Pseudomonadales; family: Moraxellaceae) were recently found to be RL producers (Rooney et al. 2009). Most other examples, however, are farther from Pseudomonas in the taxonomical class. They belong to different orders, yet under the same phylum (Gammaproteobacteria); e.g., Pseudoxanthomonas sp. (order: Xanthomonadales; family: Xanthomonadaceae; Nayak et al. 2009) and Enterobacter sp., as well as Pantoea sp. (order: Enterobacteriales; family: Enterobacteriaceae; Rooney et al. 2009; Vasileva-Tonkova and Gesheva 2007; Table 2).
Besides, RL-producing bacteria that do not belong to the class Gammaproteobacteria have been reported. The most prominent examples are various RL-producing Burkholderia spp. which are Betaproteobacteria (Andrä et al. 2006; Dubeau et al. 2009; Häussler et al. 1998; Pajarron et al. 1993). Interestingly, a Myxococcus sp. (class Deltaproteobacteria) has been reported to produce an unusual rhamnose-containing glycolipid (rhamnoside) called myxotyrosides which have a tyrosine-derived core structure glycosylated with rhamnose and acylated with unusual fatty acids such as (Z)-15-methyl-2-hexadecenoic and (Z)-2-hexadecenoic acid. Although these are not typical RLs, the authors claimed that the only molecules related to myxotyrosides are RLs and similarly acylated amino acids (Ohlendorf et al. 2008; Table 2).
Astonishingly, bacterial isolates belonging to different phyla than that of Pseudomonas, for instance, Renibacterium salmoninarum (Christova et al. 2004), Cellulomonas cellulans (Arino et al. 1998b), and Nocardioides sp. (Vasileva-Tonkova and Gesheva 2005), have been reported to produce RLs, yet they belong to the phylum Actinobacteria. Another example is Tetragenococcus koreensis, which belongs to phylum Firmicutes and has been reported to be a RL producer (Lee et al. 2005; Table 2). The glycolipid produced by C. cellulans is not a typical RL. Although not fully structurally elucidated, preliminary characterizations showed it is a glycolipid whose lipid part is composed of fatty acids and hydroxy fatty acids ranging from C10 to C18. The glycidic moiety is composed of glucose, rhamnose, and to a lesser extent, ribose (Arino et al. 1998b). It remains to be seen whether this glycolipid has surfactant activity.
Although bacterial species other than P. aeruginosa have been reported as RL producers (Table 2), it is important to note that many have not been conclusively identified (e.g., through 16S ribosomal RNA gene sequencing) nor their production of RLs have been confirmed using high-precision analytical methods (e.g., HPLC-MS). Worthy to be mentioned here, in the context of the interest in finding nonpathogenic RL-producing species for commercial production purposes, is that the pathogenicity of many of these rare, newly discovered RL-producers is often uncertain and will require further investigations. These novel isolates, reported to be RL producers, have been found distributed within and outside the class Gammaproteobacteria. This highlights the need for wide spectrum screening programs for RL producers that encompass different orders of Gammaproteobacteria or even of other classes. Still, some authors anticipate that novel RL-producing isolates will be found mostly within the class Gammaproteobacteria (Rooney et al. 2009). Finding new RL-producing bacteria is beneficial from a biotechnological point of view as it might result into the discovery of producers that are nonpathogenic compared to the pathogenic P. aeruginosa strains and hence become more appropriate candidates for the industrially-safe production of RLs.
Genetic basis of diversity of composition of natural RL mixtures
Naturally produced RL biosurfactants are always found as mixtures of different RL congeners, as observed with the various strains of P. aeruginosa (Abalos et al. 2001; Abdel-Mawgoud et al. 2009; Benincasa et al. 2004; Haba et al. 2003a; Mata-Sandoval et al. 1999; Pornsunthorntawee et al. 2008), of P. chlororaphis (Gunther et al. 2005, 2006) and of different Burkholderia spp. such as B. glumae (Pajarron et al. 1993), B. plantarii (Andrä et al. 2006), B. thailandensis, and B. pseudomallei (Dubeau et al. 2009). Variations in the nature and distribution of the different RL congeners identified in various RL mixtures might be attributed to diverse cultivation conditions as well as to strain-related variations (Déziel et al. 1999, 2000; Dubeau et al. 2009). These mixtures of congeners display physicochemical properties which together would probably be very different from the individual RL congeners.
On the genetic level, biosynthesis of RLs in P. aeruginosa occurs through three sequential steps (Soberón-Chávez et al. 2005): (1) RhlA (encoded by rhlA gene) is involved in the synthesis of the fatty acid dimer (HAAs) moiety of RLs from 3-hydroxyfatty acid precursors (Déziel et al. 2003; Lépine et al. 2002; Zhu and Rock 2008); (2) the membrane-bound RhlB rhamnosyltransferase (encoded by rhlB gene) uses dTDP-l-rhamnose and a HAA molecule as precursors, yielding mono-RL (Ochsner et al. 1994); and (3) these mono-RLs are in turn the substrates, together with dTDP-l-rhamnose, of the RhlC rhamnosyltransferase (encoded by rhlC gene) to produce di-RLs (Rahim et al. 2001). In P. aeruginosa, a bicistronic rhlAB operon encodes the first two enzymes while rhlC is elsewhere on the genome. While P. aeruginosa produces a mixture of mono- and di-RLs, P. chlororaphis has been reported to produce only mono-RLs. It was suggested that this species lack a homologue of the rhlC gene which encodes for the second rhamnosyltransferase responsible for biosynthesis of di-RLs (Gunther et al. 2005).
The genes responsible for RL synthesis in B. thailandensis and B. pseudomallei were recently reported (Dubeau et al. 2009). These bacterial species harbor two paralogous rhl gene clusters carrying homologues of rhlA, rhlB, and rhlC. However, unlike in P. aeruginosa, these three genes are grouped together in the same gene cluster. Using rhlA1, rhlA2, and rhlA1rhlA2 mutants, both rhl gene cluster paralogues were shown to contribute to the total RL production of B. thailandensis (Dubeau et al. 2009). While until then only di-RL congeners had been found to be produced by Burkholderia species, Dubeau et al. demonstrated the co-production of mono-RL congeners together with the di-RLs by B. thailandensis, although the former congeners were produced at in minimal concentrations (Dubeau et al. 2009). The ratio of di-RLs to mono-RLs found in B. thailandensis is approximately 13:1, whereas that in P. aeruginosa is only 4:1 (Dubeau et al. 2009). It is hypothesized that since P. aeruginosa harbors rhlA and rhlB in one operon and rhlC in another, this may result in different levels and timing of expression of the first and second rhamnosyltransferases (RhlB and RhlC, respectively), thus decoupling both activities. On the other hand, Burkholderia species co-express rhlB and rhlC, which might favor the simultaneous production of all the enzymes of the biosynthetic pathway in the same stoichiometric ratio. In other words, RhlC will be produced at the same time and in an amount equivalent to that of RhlB and thus would efficiently add another rhamnosyl group on most of the mono-RLs produced by RhlB.
Diversity of physiological functions and roles
Although numerous studies have been performed in the field of RLs, the exact physiological functions of these molecules for the producing bacteria are still not precisely defined. Many of these functions are derived from the well-known physicochemical properties of RLs such as surface activity, wetting ability, detergency, and other amphipathic-related properties. Taken together, studies suggest that these molecules might play multifunctional roles for the producing organisms.
RLs promote the uptake and biodegradation of poorly soluble substrates
Most bacteria degrading hydrophobic substrates release biosurfactants that facilitate the uptake and assimilation of hydrocarbons (Hommel 1994). For instance, linear alkanes are very insoluble in water, their solubility decrease as the molecular weight increases, but they are very good nutrient sources for P. aeruginosa. With the exception of n-alkanes of very low molecular weight, solubility values of these molecules are well below the micromolar range (e.g., 2 × 10−10 M for hexadecane). This impedes their sufficient uptake by bacteria, and it is thus not obvious how these hydrophobic molecules enter the cells to sustain growth. For medium- and long-chain-length n-alkanes, it is likely that bacteria gain access to them either by a surfactant-facilitated process (emulsification and solubilisation) or by directly adhering to hydrocarbon droplets (surfactant-mediated increase of cell surface hydrophobicity).
Because of their surface tension-reducing and emulsifying activities, RLs have been considered as surfactants. As a result, these amphiphilic molecules have been mostly studied for their ability to mediate the assimilation of hydrophobic substrates in liquid cultures, especially hydrocarbons such as n-alkanes (Beal and Betts 2000; Hisatsuka et al. 1971; Itoh and Suzuki 1972; Koch et al. 1991; Noordman and Janssen 2002; Shreve et al. 1995). Interestingly, RLs also enhance the biodegradation of poorly soluble molecules by causing the cell surface to become more hydrophobic (Al-Tahhan et al. 2000; Shreve et al. 1995; Zhang and Miller 1994). The few investigations performed with RL-nonproducing mutants are especially revealing: Beal and Betts (2000), using an rhlA mutant, have shown that endogenous production of RLs indeed enhances the biodegradation of hexadecane by P. aeruginosa, but that their contribution is relatively limited. In support of the notion that the specific function of RLs is unlikely to be solely to facilitate the assimilation of insoluble substrates, these exoproducts are as efficiently synthesized when the cells are grown on soluble substrates such as carbohydrates (Guerra-Santos et al. 1984).
RLs as immune modulators and virulence factors
The airway mucosa is an efficient barrier to protect the host from infections. Nevertheless, presence of noninvasive microorganisms in the airways can lead to damage to the respiratory mucosa due to the production of extracellular toxins. For instance, P. aeruginosa causes serious infections in immunocompromised patients and individuals suffering from cystic fibrosis (CF). Probably because they do not present the profile of typical or traditional virulence factors, RLs are generally not considered significant members of the arsenal of P. aeruginosa. However, a resurgence in recent literature is bringing back to the forefront some older investigations, which, together, strongly demonstrate their relevance as virulence determinants.
Kownatzki et al. (1987) found that sputum samples obtained from P. aeruginosa-colonized CF patients contained RLs (up to 8 μg/ml) and reported a correlation between elevated levels of RLs and worsened patient's clinical status. These RL concentrations in sputum samples, however, might actually underestimate the concentrations present in the lower respiratory tract, as levels of 65 μg/ml were found in secretions of a lung removed from a CF patient (Read et al. 1992).
P. aeruginosa had been known for a long time to produce a solvent-soluble glycolipid with hemolytic activity when, a few years after RLs were reported, the coincidence between the two was noticed (Sierra 1960). Nevertheless, the heat-stable extracellular hemolysin designation for RLs has persisted for many years (Fujita et al. 1988; Johnson and Boese-Marrazzo 1980; Kurioka and Liu 1967; Stutts et al. 1986). The di-RL Rha-Rha-C14-C14 from B. pseudomallei is similarly hemolytic for various erythrocyte species and also cytotoxic at high concentrations for nonphagocytic and phagocytic cell lines (Häussler et al. 1998).
Actually, purified RLs act directly on immune cells. Among many exoproducts released by P. aeruginosa that affects neutrophil recruitment, RLs have been shown to induce direct neutrophil chemotactic activity (Kharazmi et al. 1989). Preincubation of monocytes with RLs enhanced the oxidative burst response of these cells (Kharazmi et al. 1989). RLs, especially di-RLs, are cytolytic for human monocyte-derived macrophages (McClure and Schiller 1992), and at lower concentrations, they can also inhibit the phagocytic response of macrophages (McClure and Schiller 1996). Tissue-cultured macrophages treated with the B. pseudomallei di-RLs internalized fewer viable B. pseudomallei bacteria (Häussler et al. 2003). It is worth mentioning here that RLs are known to induce lysis of the amoebae Dictyostelium discoideum (Cosson et al. 2002). Rha-Rha-C14-C14 is also produced by B. plantarii and was found to be endowed with endotoxin-like properties (Andrä et al. 2006). P. aeruginosa RLs induce histamine release from mast cells (Bergmann et al. 1989) and stimulate the generation and release of the inflammatory mediators serotonin and 12-hydroxyeicosatetraenoic acid from human platelets (Konig et al. 1992). RLs stimulate the copious release of interleukin (IL)-8, granulocyte-macrophage colony-stimulating factor, and IL-6 from nasal epithelial cells at non-cytotoxic levels (Bédard et al. 1993). RLs induce the lysis of polymorphonuclear leukocytes (PMNs; Jensen et al. 2007; Shryock et al. 1984; Van Gennip et al. 2009) while, at subtoxic levels, they only stimulate both chemotaxis and chemokinesis of PMNs (Shryock et al. 1984). Furthermore, Alhede et al. (2009) recently demonstrated that exposure to PMNs triggers the production of RLs by biofilm-growing P. aeruginosa cells, whereas in vitro, their biofilms usually produce very little RLs (Alhede et al. 2009; Morici et al. 2007), suggesting that biofilm-associated RLs function as a shield against PMNs. These studies illustrate that RLs probably contribute to the inflammatory-related tissue damage observed in lungs of CF patient.
RLs inhibit ciliary function and produce damage to the bronchial epithelium: they alter respiratory epithelial ion transport through reduced sodium absorption and unidirectional chloride fluxes across human bronchial epithelium (Stutts et al. 1986), inhibit transcellular ion transport, as observed by a decrease in amiloride-sensitive short-circuit current across sheep tracheal epithelium (Graham et al. 1993), interfere with the normal tracheal ciliary function, as shown on ciliary function in rabbit tracheal epithelium (Hingley et al. 1986), and confirmed by a slowing of ciliary beat frequency of human airway epithelium in vitro (Kanthakumar et al. 1996; Read et al. 1992). They also stimulate the release of mucus glycoconjugates from feline trachea or human bronchial mucosa (Fung et al. 1995; Somerville et al. 1992). More recently, Zulianello et al. (2006) have shown that P. aeruginosa requires the production of RLs to invade respiratory epithelia reconstituted with primary human respiratory cells.
Finally, the importance of RL production for establishment of an actual infection was recently directly shown with two in vivo models: compared to the wild type strain, an rhlA mutant was more rapidly cleared from both mice models of intraperitoneal foreign-body infection and pulmonary infection (Van Gennip et al. 2009).
RLs as antimicrobials
Early on, the wide-ranging antimicrobial properties of RLs were noted (Itoh et al. 1971). Especially, they were shown to be active against a large variety of bacteria, including both Gram-negative and Gram-positive species, though the latter, such as Bacillus subtilis, are in general much more susceptible to the toxic effect of RLs (Itoh et al. 1971; Lang et al. 1989). Growth inhibition was also observed against Rhodococcus erythropolis and Bacillus cereus (Arino et al. 1998a).
The target of synthetic surfactants is the cell envelope. Likewise, the proposed mechanism of action of RLs consists of intercalation into the biological membrane and destruction by their permeabilizing effect (Sotirova et al. 2008).
The group of Manresa has performed comprehensive investigations on the antimicrobial properties of mixtures of RL congeners produced by three different strains of P. aeruginosa grown on various types of vegetable oil wastes (Abalos et al. 2001; Benincasa et al. 2004; Haba et al. 2003b). The various RL combinations displayed antimicrobial activity against nearly all tested Gram-positives species, including Staphylococcus, Mycobacterium, and Bacillus, and significant activity against a number of Gram-negative species, with Serratia marcescens, Enterobacter aerogenes, and Klebsiella pneumoniae as the especially sensitive ones. They also found an excellent inhibitory activity against a range of fungal species, including the filamentous fungi Chaetomium globosum, Aureobacidium pullulans, and Gliocladium virens, and the phytopathogens Botrytis cinerea and Rhizoctonia solanii, but no significant effect on yeasts (Abalos et al. 2001; Benincasa et al. 2004; Haba et al. 2003b).
RLs were shown to exert high zoosporicidal activity, probably through zoospore lysis, against various zoosporic phytopathogens, including species from the Pythium, Phytophthora, and Plasmopara genera (Stanghellini and Miller 1997). Interestingly, fluorescent pseudomonads are effective in the biological control of plant pathogens. Recently, the biocontrol activity of P. aeruginosa strain PNA1 was reported to require the production of RLs, as an rhlA mutant was much less successful at inhibiting plant disease caused by Pythium sp. (Perneel et al. 2008).
Furthermore, antiviral, algicidal, mycoplasmicidal, and antiamoebal properties of RLs have also been reported (Cosson et al. 2002; Haferburg et al. 1987; Itoh et al. 1971; Wang et al. 2005).
RLs in surface motility
Unlike swimming motility, which bacteria use to move individually in the aqueous phase, swarming motility is a form of rapid and coordinated translocation of a bacterial population across a surface, often resulting in flowery, dendritic colonies (Fraser and Hughes 1999; Verstraeten et al. 2008). Swarmer cells move along their long axis in multicellular rafts, are often hyperflagellated and elongated, and are surrounded by a self-produced “slime” mainly composed of exopolysaccharides and surface-active compounds (Harshey 2003). A key feature of swarming cells is surface conditioning achieved by the production of a wetting agent that reduces the surface tension surrounding the colony (Matsuyama and Nakagawa 1996).
In P. aeruginosa, swarming motility was relatively recently reported (Déziel et al. 2001; Köhler et al. 2000; Rashid and Kornberg 2000) and shown to require flagella and the production of a wetting agent, at least HAAs, and ideally also RLs (Caiazza et al. 2005; Déziel et al. 2003; Köhler et al. 2000). A notable exception was noted by Shrout et al. (2006) who were able to see swarming of an rhlAB mutant. Recently, Tremblay et al. (2007) reported that HAAs and di-RLs actually modulate the development of the fractal-like patterns formed by migrating P. aeruginosa swarming colonies. They found that di-RLs promote tendril formation and migration, acting like self-produced chemotactic attractants, while HAAs play the opposite role, repelling swarming tendrils. Mono-RLs seem to act solely as wetting agent. These findings extended the previous work by Caiazza et al. (2005) who identified a role for RLs in maintenance of the swarming colony organization.
An rhlA mutant was also reported to display reduced twitching motility, a form of surface translocation requiring type IV pili but not flagella (Pamp and Tolker-Nielsen 2007). Finally, under the exact same conditions of swarming, P. aeruginosa expressing neither flagella nor type IV pili displays another type of motility called sliding, which was also shown to be promoted by the production of RLs (Murray and Kazmierczak 2008).
RLs in biofilm development
More and more studies are finding that certain factors which affect swarming motility also influence other bacterial multicellular behaviors, especially biofilm formation (Harshey 2003; Verstraeten et al. 2008). P. aeruginosa mutants with altered swarming are defective in “normal,” mature biofilm formation (Overhage et al. 2007; Shrout et al. 2006; Yeung et al. 2009). Indeed, mutants displaying enhanced swarming are much less prone to form a biofilm (Caiazza et al. 2007; Parkins et al. 2001), presumably because the cells do not efficiently settle on the substratum. Accumulating data therefore support the model that swarming motility is required as a critical step in biofilm development.
One explanation for the fundamental link between swarming motility and biofilms in P. aeruginosa is being revealed: RLs are also involved in biofilm development. First, exogenous RLs induce a release of lipopolysaccharides and thus enhance cell surface hydrophobicity, which might favor primary adhesion of planktonic cells (Al-Tahhan et al. 2000; Zhang and Miller 1994). RL production appears as a means for regulation of cell-surface hydrophobicity and modification of adhesive interactions, especially when changes in nutritional conditions occur (Boles et al. 2005; de Andrès et al. 1991; Déziel et al. 2003; Neu 1996).
RLs facilitate the surface-associated migration of bacteria in the biofilm and therefore the initial microcolony formation and differentiation of the biofilm structure (Pamp and Tolker-Nielsen 2007). Using fluorescent reporter genes, Lequette and Greenberg (2005) have shown that the expression of rhlAB is modulated by the localization of cells in the biofilm. Actually, a critical role for RLs seems to involve interference with both cell–cell and cell–substratum interactions, as they were shown to mediate the detachment and dispersion of P. aeruginosa cells from the biofilm, especially from the center of microcolonies (Boles et al. 2005; Schooling et al. 2004) and to maintain open the channels inside the biofilm during late stages of biofilm development (Davey et al. 2003). Accordingly, overproduction of RLs by P. aeruginosa, as observed for instance in algR mutant biofilms (Morici et al. 2007), disrupts the structure or impede biofilm formation (Davey et al. 2003). Exogenous RLs interfere with normal biofilm infrastructure, and this detachment effect seems nonspecific, as addition of RLs disrupts Salmonella typhimurium and Bordetella bronchiseptica biofilms as well (Irie et al. 2005; Mireles et al. 2001).
Perspectives
In this mini-review, we have presented a detailed assessment of the diversity of RL structures, RL-producing microorganisms, and physiological functions of RLs for these producing bacteria. Clearly, the widening variety of microbes identified as RL producers will entice researchers to screen for nonpathogenic and industrially safe alternatives to P. aeruginosa, the well-known opportunistic human pathogen. However, it must be emphasized that in most cases when a new bacterial species was reported to produce RLs, often only one isolate was actually selected for further studies. Therefore, for now, only few bacterial species can be considered actual RL producers. Screening efforts will hence require the development of wide spectrum approaches, probably based on the detection of surface tension lowering substance or, theoretically and ideally, the presence of the rhlA, rhlB, and rhlC homologues in the genomes of bacteria.
Likely, there will be trials to separate the main RL homologues in pure forms to explore their individual biological and surface activities for a variety of applications. This approach may unravel the promising biotechnological uses of these glycolipid biosurfactants either in pure form or in mixtures. Moreover, characterization of the structure-activity relationship of these compounds might be useful for prediction of the behavior of different natural RL mixtures comprising one major congener among other minor congeners.
Finally, it is expected that studies on the genetic regulation of RL synthesis, especially in the context of specific environmental conditions affecting the requirement for the production of these biosurfactants, will help in deciphering the exact genetic control of RL production. Consequently, this understanding should lead to more successful control of regulatory and growth factors directing the optimized large-scale production with a safe host.
References
Abalos A, Pinazo A, Infante MR, Casals M, Garcia F, Manresa A (2001) Physicochemical and antimicrobial properties of new rhamnolipids produced by Pseudomonas aeruginosa AT10 from soybean oil refinery wastes. Langmuir 17:1367–1371
Abdel-Mawgoud AM, Aboulwafa MM, Hassouna NAH (2009) Characterization of rhamnolipid produced by Pseudomonas aeruginosa isolate Bs20. Appl Biochem Biotechnol 157:329–345
Abouseoud M, Maachi R, Amrane A, Boudergua S, Nabi A (2008a) Evaluation of different carbon and nitrogen sources in production of biosurfactant by Pseudomonas fluorescens. Conference on Desalination and the Environment. Sani Resort, Halkidiki, Greece, pp 143–151
Abouseoud M, Yataghene A, Amrane A, Maachi R (2008b) Biosurfactant production by free and alginate entrapped cells of Pseudomonas fluorescens. J Ind Microbiol Biotech 35:1303–1308
Alhede M, Bjarnsholt T, Jensen PO, Phipps RK, Moser C, Christophersen L, Christensen LD, van Gennip M, Parsek M, Hoiby N, Rasmussen TB, Givskov M (2009) Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiol Sgm 155:3500–3508
Al-Tahhan RA, Sandrin TR, Bodour AA, Maier RM (2000) Rhamnolipid-induced removal of lipopolysaccharide from Pseudomonas aeruginosa: effect on cell surface properties and interaction with hydrophobic substrates. Appl Environ Microbiol 66:3262–3268
Andrä J, Rademann J, Howe J, Koch MHJ, Heine H, Zähringer U, Brandenburg K (2006) Endotoxin-like properties of a rhamnolipid exotoxin from Burkholderia (Pseudomonas) plantarii: immune cell stimulation and biophysical characterization. Biol Chem 387:301–310
Arino S, Marchal R, Vandecasteele JP (1996) Identification and production of a rhamnolipidic biosurfactant by a Pseudomonas species. Appl Microbiol Biotechnol 45:162–168
Arino S, Marchal R, Vandecasteele JP (1998a) Involvement of a rhamnolipid-producing strain of Pseudomonas aeruginosa in the degradation of polycyclic aromatic hydrocarbons by a bacterial community. J Appl Microbiol 84:769–776
Arino S, Marchal R, Vandecasteele JP (1998b) Production of new extracellular glycolipids by a strain of Cellulomonas cellulans (Oerskovia xanthineolytica) and their structural characterization. Can J Microbiol 44:238–243
Bauer J, Brandenburg K, Zahringer U, Rademann J (2006) Chemical synthesis of a glycolipid library by a solid-phase strategy allows elucidation of the structural specificity of immunostimulation by rhamnolipids. Chem Eur J 12:7116–7124
Beal R, Betts WB (2000) Role of rhamnolipid biosurfactants in the uptake and mineralization of hexadecane in Pseudomonas aeruginosa. J Appl Microbiol 89:158–168
Bédard M, McClure CD, Schiller NL, Francoeur C, Cantin A, Denis M (1993) Release of interleukin-8, interleukin-6, and colony-stimulating factors by upper airway epithelial cells: implication for cystic fibrosis. Am J Resir Cell Mol Biol 9:455–462
Benincasa M, Abalos A, Oliveira I, Manresa A (2004) Chemical structure, surface properties and biological activities of the biosurfactant produced by Pseudomonas aeruginosa LBI from soapstock. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol 85:1–8
Bergmann U, Scheffer J, Koller M, Schonfeld W, Erbs G, Muller FE, Konig W (1989) Induction of inflammatory mediators (histamine and leukotrienes) from rat peritoneal mast cells and human granulocytes by Pseudomonas aeruginosa strains from burn patients. Infect Immun 57:2187–2195
Bergström S, Theorell H, Davide H (1946a) On a metabolic product of Ps. pyocyanea. Pyolipic acid, active against M. tuberculosis. Arkiv Chem Mineral Geol 23A(13):1–12
Bergström S, Theorell H, Davide H (1946b) Pyolipic acid. A metabolic product of Pseudomonas pyocyanea active against Mycobacterium tuberculosis. Arch Biochem Biophys 10:165–166
Boles BR, Thoendel M, Singh PK (2005) Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol 57:1210–1223
Burger MM, Glaser L, Burton RM (1963) The enzymatic synthesis of a rhamnose-containing glycolipid by extracts of Pseudomonas aeruginosa. J Biol Chem 238:2595–2602
Caiazza NC, Shanks RMQ, O'Toole GA (2005) Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361
Caiazza NC, Merritt JH, Brothers KM, O'Toole GA (2007) Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:3603–3612
Celik GY, Aslim B, Beyatli Y (2008) Enhanced crude oil biodegradation and rhamnolipid production by Pseudomonas stutzeri strain G11 in the presence of Tween-80 and Triton X-100. J Environ Biol 29:867–870
Christova N, Tuleva B, Lalchev Z, Jordanova A, Jordanov B (2004) Rhamnolipid biosurfactants produced by Renibacterium salmoninarum 27BN during growth on n-hexadecane. Z Nat Forsch C J Biosci 59:70–74
Cosson P, Zulianello L, Join-Lambert O, Faurisson F, Gebbie L, Benghezal M, van Delden C, Curty LK, Köhler T (2002) Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J Bacteriol 184:3027–3033
Cuny P, Acquaviva M, Gilewicz M (2004) Phenanthrene degradation, emulsification and surface tension activities of a Pseudomonas putida strain isolated from a coastal oil contaminated microbial mat. Ophelia 58:283–287
Davey ME, Caiazza NC, O'Toole GA (2003) Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185:1027–1036
de Andrès C, Espuny MJ, Robert M, Mercadé ME, Guinea J (1991) Cellular lipid accumulation by Pseudomonas aeruginosa 44T1. Appl Microbiol Biotechnol 35:813–816
Desai JD, Banat IM (1997) Microbial production of surfactants and their commercial potential. Microbiol Mol Biol Rev 61:47–64
Déziel E, Lépine F, Dennie D, Boismenu D, Mamer OA, Villemur R (1999) Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim Biophys Acta Mol Cell Biol Lipids 1440:244–252
Déziel E, Lépine F, Milot S, Villemur R (2000) Mass spectrometry monitoring of rhamnolipids from a growing culture of Pseudomonas aeruginosa strain 57RP. Biochim Biophys Acta Mol Cell Biol Lipids 1485:145–152
Déziel E, Comeau Y, Villemur R (2001) Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol 183:1195–1204
Déziel E, Lépine F, Milot S, Villemur R (2003) rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiol Sgm 149:2005–2013
Dubeau D, Déziel E, Woods D, Lépine F (2009) Burkholderia thailandensis harbors two identical rhl gene clusters responsible for the biosynthesis of rhamnolipids. BMC Microbiology 9:263
Edwards JR, Hayashi JA (1965) Structure of a rhamnolipid from Pseudomonas aeruginosa. Arch Biochem Biophys 111:415–421
Fraser GM, Hughes C (1999) Swarming motility. Curr Opin Microbiol 2:630–635
Fujita K, Akino T, Yoshioka H (1988) Characteristics of the heat-stable extracellular hemolysin from Pseudomonas aeruginosa. Infect Immun 56:1385–1387
Fung DC, Somerville M, Richardson PS, Sheehan JK (1995) Mucus glycoconjugate complexes released from feline trachea by a bacterial toxin. Am J Respir Cell Mol Biol 12:296–306
Graham A, Steel DM, Wilson R, Cole PJ, Alton E, Geddes DM (1993) Effects of purified Pseudomonas rhamnolipids on bioelectric properties of sheep tracheal epithelium. Exp Lung Res 19:77–89
Gruber T, Chmiel H, Käppeli O, Sticher P, Fiechter A (1993) Integrated process for continuous rhamnolipid biosynthesis. In: Kosaric N (ed) Surfactant science series—biosurfactants: production, properties, application. Marcel Dekker, Inc, New York, pp 157–173
Guerra-Santos L, Kappeli O, Fiechter A (1984) Pseudomonas aeruginosa biosurfactant production in continuous culture with glucose as carbon source. Appl Environ Microbiol 48:301–305
Gunther NW, Nunez A, Fett W, Solaiman DKY (2005) Production of rhamnolipids by Pseudomonas chlororaphis, a nonpathogenic bacterium. Appl Environ Microbiol 71:2288–2293
Gunther NW, Nunez A, Fortis L, Solaiman DKY (2006) Proteomic based investigation of rhamnolipid production by Pseudomonas chlororaphis strain NRRL B-30761. J Ind Microbiol Biotech 33:914–920
Guo YP, Hu YY, Gu RR, Lin H (2009) Characterization and micellization of rhamnolipidic fractions and crude extracts produced by Pseudomonas aeruginosa mutant MIG-N146. J Colloid Interface Sci 331:356–363
Haba E, Abalos A, Jauregui O, Espuny MJ, Manresa A (2003a) Use of liquid chromatography-mass spectroscopy for studying the composition and properties of rhamnolipids produced by different strains of Pseudomonas aeruginosa. J Surfactants Deterg 6:155–161
Haba E, Pinazo A, Jauregui O, Espuny MJ, Infante MR, Manresa A (2003b) Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol Bioeng 81:316–322
Haferburg D, Hommel R, Kleber HP, Kluge S, Schuster G, Zschiegner HJ (1987) Antiphytovirale Aktivität von Rhamnolipid aus Pseudomonas aeruginosa. Acta Biotechnol 7:353–356
Harshey RM (2003) Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273
Hauser G, Karnovsky ML (1954) Studies on the production of glycolipide by Pseudomonas aeruginosa. J Bacteriol 68:645–654
Häussler S, Nimtz M, Domke T, Wray V, Steinmetz I (1998) Purification and characterization of a cytotoxic exolipid of Burkholderia pseudomallei. Infect Immun 66:1588–1593
Häussler S, Rohde M, von Neuhoff N, Nimtz M, Steinmetz I (2003) Structural and functional cellular changes induced by Burkholderia pseudomallei rhamnolipid. Infect Immun 71:2970–2975
Hingley ST, Hastie A, Kueppers F, Higgins ML, Weinbaum G, Shryock T (1986) Effect of ciliostatic factors from Pseudomonas aeruginosa on rabbit respiratory cilia. Infect Immun 51:254–262
Hirayama T, Kato I (1982) Novel methyl rhamnolipids from Pseudomonas aeruginosa. FEBS Lett 139:81–85
Hisatsuka K-I, Nakahara T, Sang N, Yamada K (1971) Formation of rhamnolipid by Pseudomonas aeruginosa and its function in hydrocarbon fermentation. Agric Biol Chem 35:686–692
Hommel R (1994) Formation and function of biosurfactants for degradation of water-insoluble substrates. In: Ratledge C (ed) Biochemistry of microbial degradation. Kluwer Academic Publishers, London, pp 63–87
Howe J, Bauer J, Andra J, Schromm AB, Ernst M, Rossle M, Zahringer U, Rademann J, Brandenburg K (2006) Biophysical characterization of synthetic rhamnolipids. Febs Journal 273:5101–5112
Husain S (2008) Effect of surfactants on pyrene degradation by Pseudomonas fluorescens 29L. World J Microbiol Biotechnol 24:2411–2419
Irie Y, O'Toole GA, Yuk MH (2005) Pseudomonas aeruginosa rhamnolipids disperse Bordetella bronchiseptica biofilms. Fems Microbiol Lett 250:237–243
Ishigami Y, Gama Y, Nagahora H, Yamaguchi M, Nakahara H, Kamata T (1987a) The pH-sensitive conversion of molecular aggregates of rhamnolipid biosurfactant. Chem Lett 16:763–766
Ishigami Y, Gama Y, Yamaguchi M, Nakahara H, Kamata T (1987b) Surface active properties of rhamnolipids as microbial biosurfactants. J Jpn Oil ChemSoc 36:791–796
Ishigami Y, Ishii F, Choi YK, Kajiuchi T (1996) Estimation of polarity and fluidity of colloidal interfaces and biosurfaces using rhamnolipid B pyrenacylester as surface-active fluorescent probe. Colloid Surf B 7:215–220
Itoh S, Suzuki T (1972) Effect of rhamnolipids on growth of Pseudomonas aeruginosa mutant deficient in n-paraffin-utilizing ability. Agric Biol Chem 36:2233–2235
Itoh S, Honda H, Tomita F, Suzuki T (1971) Rhamnolipids produced by Pseudomonas aeruginosa grown on n-paraffin (mixture of C12, C13 and C14 fractions). J Antibiot 24:855–859
Janiyani KL, Wate SR, Joshi SR (1992) Surfactant production by Pseudomonas stutzeri. J Microb Biotechnol 7:18–21
Jarvis FG, Johnson MJ (1949) A Glyco-lipide produced by Pseudomonas aeruginosa. J Am Chem Soc 71:4124–4126
Jensen PO, Bjarnsholt T, Phipps R, Rasmussen TB, Calum H, Christoffersen L, Moser C, Williams P, Pressler T, Givskov M, Hoiby N (2007) Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiol Sgm 153:1329–1338
Johnson MK, Boese-Marrazzo D (1980) Production and properties of heat-stable extracellular hemolysin from Pseudomonas aeruginosa. Infect Immun 29:1028–1033
Kanthakumar K, Taylor GW, Cundell DR, Dowling RB, Johnson M, Cole PJ, Wilson R (1996) The effect of bacterial toxins on levels of intracellular adenosine nucleotides and human ciliary beat frequency. Pulm Pharmacol Ther 9:223–230
Kharazmi A, Bibi Z, Nielsen H, Hoiby N, Döring G (1989) Effect of Pseudomonas aeruginosa rhamnolipid on human neutrophil and monocyte function. Apmis 97:1068–1072
Koch AK, Käppeli O, Fiechter A, Reiser J (1991) Hydrocarbon assimilation and biosurfactant production in Pseudomonas aeruginosa mutants. J Bacteriol 173:4212–4219
Köhler T, Curty LK, Barja F, van Delden C, Pechère JC (2000) Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996
König B, Bergmann U, König W (1992) Induction of inflammatory mediator release (serotonin and 12- hydroxyeicosatetraenoic acid) from human platelets by Pseudomonas aeruginosa glycolipid. Infect Immun 60:3150–3155
Kownatzki R, Tümmler B, Döring G (1987) Rhamnolipid of Pseudomonas aeruginosa in sputum of cystic fibrosis patients. Lancet 1:1026–1027
Kurioka S, Liu PV (1967) Effect of the hemolysin of Pseudomonas aeruginosa on phosphatides and on phospholipase C activity. J Bacteriol 93:670–674
Lang S, Wagner F (1987) Structures and properties of biosurfactants. In: Kosaric N, Cairns WL, Gray NCC (eds) Biosurfactants and biotechnology. Marcel Dekker Inc, New York, pp 21–45
Lang S, Wullbrandt D (1999) Rhamnose lipids—biosynthesis, microbial production and application potential. Appl Microbiol Biotechnol 51:22–32
Lang S, Katsiwela E, Wagner F (1989) Antimicrobial effects of biosurfactants. Fat Sci Technol 91:363–366
Lee M, Kim MK, Vancanneyt M, Swings J, Kim SH, Kang MS, Lee ST (2005) Tetragenococcus koreensis sp. nov., a novel rhamnolipid-producing bacterium. Int J Syst Evol Microbiol 55:1409–1413
Lépine F, Déziel E, Milot S, Villemur R (2002) Liquid chromatographic/mass spectrometric detection of the 3-(3-hydroxyalkanoyloxy)alkanoic acid precursors of rhamnolipids in Pseudomonas aeruginosa cultures. J Mass Spectrom 37:41–46
Lequette Y, Greenberg EP (2005) Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J Bacteriol 187:37–44
Maier RM, Soberón-Chávez G (2000) Pseudomonas aeruginosa rhamnolipids: biosynthesis and potential applications. Appl Microbiol Biotechnol 54:625–633
Martinez-Toledo A, Rios-Leal E, Vazquez-Duhalt R, Gonzalez-Chavez Mdel C, Esparza-Garcia JF, Rodriguez-Vazquez R (2006) Role of phenanthrene in rhamnolipid production by P. putida in different media. Environ Technol 27:137–142
Mata-Sandoval JC, Karns J, Torrents A (1999) High-performance liquid chromatography method for the characterization of rhamnolipid mixtures produced by Pseudomonas aeruginosa UG2 on corn oil. J Chromatogr A 864:211–220
Matsuyama T, Nakagawa Y (1996) Surface-active exolipids: analysis of absolute chemical structures and biological functions. J Microbiol Methods 25:165–175
McClure CD, Schiller NL (1992) Effects of Pseudomonas aeruginosa rhamnolipids on monocyte-derived macrophages. J Leukoc Biol 51:97–102
McClure CD, Schiller NL (1996) Inhibition of macrophage phagocytosis by Pseudomonas aeruginosa rhamnolipids in vitro and in vivo. Curr Microbiol 33:109–117
Mireles JR 2nd, Toguchi A, Harshey RM (2001) Salmonella enterica serovar typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J Bacteriol 183:5848–5854
Morici LA, Carterson AJ, Wagner VE, Frisk A, Schurr JR, Bentrup KHZ, Hassett DJ, Iglewski BH, Sauer K, Schurr MJ (2007) Pseudomonas aeruginosa AlgR represses the Rhl quorum-sensing system in a biofilm-specific manner. J Bacteriol 189:7752–7764
Murray TS, Kazmierczak BI (2008) Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol 190:2700–2708
Nayak AS, Vijaykumar MH, Karegoudar TB (2009) Characterization of biosurfactant produced by Pseudoxanthomonas sp PNK-04 and its application in bioremediation. Int Biodeterior Biodegrad 63:73–79
Neu TR (1996) Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol Rev 60:151–166
Nitschke M, Costa S, Contiero J (2005) Rhamnolipid surfactants: an update on the general aspects of these remarkable biomolecules. Biotechnol Prog 21:1593–1600
Nitschke M, Costa S, Contiero J (2009) Structure and applications of a rhamnolipid surfactant produced in soybean oil waste. Appl Biochem Biotechnol. doi:10.1007/s12010-009-8707-8
Noordman WH, Janssen DB (2002) Rhamnolipid stimulates uptake of hydrophobic compounds by Pseudomonas aeruginosa. Appl Environ Microbiol 68:4502–4508
Ochsner UA, Fiechter A, Reiser J (1994) Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem 269:19787–19795
Ochsner UA, Hembach T, Fiechter A (1996) Production of rhamnolipid biosurfactants. Adv Biochem Eng Biotechnol 53:89–118
Ohlendorf B, Lorenzen W, Kehraus S, Krick A, Bode HB, König GM (2008) Myxotyrosides A and B, unusual rhamnosides from Myxococcus sp. J Nat Products 72:82–86
Oliveira FJS, Vazquez L, de Campos NP, de Franca FP (2009) Production of rhamnolipids by a Pseudomonas alcaligenes strain. Process Biochem 44:383–389
Onbasli D, Aslim B (2009) Biosurfactant production in sugar beet molasses by some Pseudomonas spp. J Environ Biol 30:161–163
Overhage J, Lewenza S, Marr AK, Hancock REW (2007) Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J Bacteriol 189:2164–2169
Pajarron AM, Dekoster CG, Heerma W, Schmidt M, Haverkamp J (1993) Structure identification of natural rhamnolipid mixtures by fast-atom-bombardment tandem mass-spectrometry. Glycoconj J 10:219–226
Pamp SJ, Tolker-Nielsen T (2007) Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J Bacteriol 189:2531–2539
Parkins MD, Ceri H, Storey DG (2001) Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol Microbiol 40:1215–1226
Perneel M, D'Hondt L, De Maeyer K, Adiobo A, Rabaey K, Hofte M (2008) Phenazines and biosurfactants interact in the biological control of soil-borne diseases caused by Pythium spp. Environ Microbiol 10:778–788
Pornsunthorntawee O, Wongpanit P, Chavadej S, Abe M, Rujiravanit R (2008) Structural and physicochemical characterization of crude biosurfactant produced by Pseudomonas aeruginosa SP4 isolated from petroleum-contaminated soil. Bioresour Technol 99:1589–1595
Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberón-Chávez G (2001) Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol Microbiol 40:708–718
Rahman P, Lungut A, Idowu J, Olea M (2009) Biosurfactant production using novel bacteria from Northeast England. Building business on bioscience sustainable innovation conference. Heriot-Watt University, Edinburgh
Rashid MH, Kornberg A (2000) Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 97:4885–4890
Read RC, Roberts P, Munro N, Rutman A, Hastie A, Shryock T, Hall R, McDonald-Gibson W, Lund V, Taylor G (1992) Effect of Pseudomonas aeruginosa rhamnolipids on mucociliary transport and ciliary beating. J Appl Physiol 72:2271–2277
Rendell NB, Taylor GW, Somerville M, Todd H, Wilson R, Cole PJ (1990) Characterization Of Pseudomonas Rhamnolipids. Biochim Biophys Acta 1045:189–193
Rooney AP, Price NP, Ray KJ, Kuo TM (2009) Isolation and characterization of rhamnolipid-producing bacterial strains from a biodiesel facility. FEMS Microbiol Lett 295:82–87
Schenk T, Breitschwerdt A, Kessels G, Schuphan I, Schmidt B (1997) A biosynthetic route to [C-14]-labelled rhamnolipids. J Label Comp Radiopharm 39:705–710
Schooling SR, Charaf UK, Allison DG, Gilbert P (2004) A role for rhamnolipid in biofilm dispersion. Biofilms 1:91–99
Sharma A, Jansen R, Nimtz M, Johri BN, Wray V (2007) Rhamnolipids from the rhizosphere bacterium Pseudomonas sp. GRP(3) that reduces damping-off disease in Chilli and tomato nurseries. J Nat Prod 70:941–947
Shaw N (1970) Bacterial glycolipids. Microbiol Mol Biol Rev 34:365–377
Shreve GS, Inguva S, Gunnam S (1995) Rhamnolipid biosurfactant enhancement of hexadecane biodegradation by Pseudomonas aeruginosa. Mol Mar Biol Biotechnol 4:331–337
Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR (2006) The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol 62:1264–1277
Shryock TR, Silver SA, Banschbach MW, Kramer JC (1984) Effect of Pseudomonas aeruginosa rhamnolipid on human neutrophil migration. Curr Microbiol 10:323–328
Sierra G (1960) Hemolytic effect of a glycolipid produced by Pseudomonas aeruginosa. Antonie Van Leeuwenhoek 26:189–192
Soberón-Chávez G (2004) Biosynthesis of rhamnolipids. In: Ramos JL (ed) Pseudomonas. Kluwer Academic/Plenum Publishers, New York, pp 173–189
Soberón-Chávez G, Lépine F, Déziel E (2005) Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol 68:718–725
Somerville M, Taylor G, Watson D, Rendell NB, Rutman A, Todd H, Davies JR, Wilson R, Cole P, Richardson PS (1992) Release of mucus glycoconjugates by Pseudomonas aeruginosa rhamnolipid into feline trachea in vivo and human bronchus in vitro. Am J Respir Cell Mol Biol 6:116–122
Sotirova AV, Spasova DI, Galabova DN, Karpenko E, Shulga A (2008) Rhamnolipid-biosurfactant permeabilizing effects on gram-positive and gram-negative bacterial strains. Curr Microbiol 56:639–644
Stanghellini ME, Miller RM (1997) Biosurfactants: their identity and potential efficacy in the biological control of zoosporic plant pathogens. Plant Dis 81:4–12
Stutts MJ, Schwab JH, Chen MG, Knowles MR, Boucher RC (1986) Effects of Pseudomonas aeruginosa on bronchial epithelial ion transport. Am Rev Respir Dis 134:17–21
Syldatk C, Lang S, Wagner F, Wray V, Witte L (1985) Chemical and physical characterization of four interfacial-active rhamnolipids from Pseudomonas spec. DSM 2874 grown on n-alkanes. Z Naturforsch C 40:51–60
Tremblay J, Richardson AP, Lépine F, Déziel E (2007) Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ Microbiol 9:2622–2630
Tuleva BK, Ivanov GR, Christova NE (2002) Biosurfactant production by a new Pseudomonas putida strain. Z Nat Forsch C J Biosci 57:356–360
Urakami T, Ito-Yoshida C, Araki H, Kijima T, Suzuki KI, Komagata K (1994) Transfer of Pseudomonas plantarii and Pseudomonas glumae to Burkholderia as Burkholderia spp. and description of Burkholderia vandii sp. nov. Int J Syst Bacteriol 44:235–245
Van Gennip M, Christensen LD, Alhede M, Phipps R, Jensen PO, Christophersen L, Pamp SJ, Moser C, Mikkelsen PJ, Koh AY, Tolker-Nielsen T, Pier GB, Hoiby N, Givskov M, Bjarnsholt T (2009) Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. Apmis 117:537–546
Vasileva-Tonkova E, Gesheva V (2005) Glycolipids produced by Antarctic Nocardioides sp during growth on n-paraffin. Process Biochem 40:2387–2391
Vasileva-Tonkova E, Gesheva V (2007) Biosurfactant production by antarctic facultative anaerobe Pantoea sp during growth on hydrocarbons. Curr Microbiol 54:136–141
Vasileva-Tonkova E, Galabova D, Stoimenova E, Lalchev Z (2006) Production and properties of biosurfactants from a newly isolated Pseudomonas fluorescens HW-6 growing on hexadecane. Z Nat Forsch C J Biosci 61:553–559
Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J (2008) Living on a surface: swarming and biofilm formation. Trends Microbiol 16:496–506
Wang XL, Gong LY, Liang SK, Han XR, Zhu CJ, Li YB (2005) Algicidal activity of rhamnolipid biosurfactants produced by Pseudomonas aeruginosa. Harmful Algae 4:433–443
Wilson NG, Bradley G (1996) The effect of immobilization on rhamnolipid production by Pseudomonas fluorescens. J Appl Bacteriol 81:525–530
Yamaguchi M, Sato M, Yamada K (1976) Microbial production of sugar lipids. Chem Ind 17:741–742
Yamaguchi M, Sato A, Dazai M, Takahara Y (1978) Report of Ferment. Res Inst 51:51
Yeung AT, Torfs EC, Jamshidi F, Bains M, Wiegand I, Hancock RE, Overhage J (2009) Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J Bacteriol 191:5592–5602
Zhang YM, Miller RM (1994) Effect of a Pseudomonas rhamnolipid biosurfactant on cell hydrophobicity and biodegradation of octadecane. Appl Environ Microbiol 60:2101–2106
Zhu K, Rock CO (2008) RhlA converts beta-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the beta-hydroxydecanoyl-beta-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J Bacteriol 190:3147–3154
Zulianello L, Canard C, Köhler T, Caille D, Lacroix JS, Meda P (2006) Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect Immun 74:3134–3147
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Abdel-Mawgoud, A.M., Lépine, F. & Déziel, E. Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol 86, 1323–1336 (2010). https://doi.org/10.1007/s00253-010-2498-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2498-2