Abstract
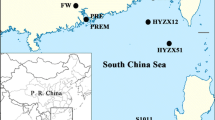
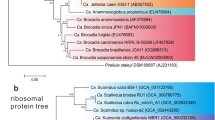
Published polymerase chain reaction primer sets for detecting the genes encoding 16S rRNA gene and hydrazine oxidoreductase (hzo) in anammox bacteria were compared by using the same coastal marine sediment samples. While four previously reported primer sets developed to detect the 16S rRNA gene showed varying specificities between 12% and 77%, an optimized primer combination resulted in up to 98% specificity, and the recovered anammox 16S rRNA gene sequences were >95% sequence identical to published sequences from anammox bacteria in the Candidatus “Scalindua” group. Furthermore, four primer sets used in detecting the hzo gene of anammox bacteria were highly specific (up to 92%) and efficient, and the newly designed primer set in this study amplified longer hzo gene segments suitable for phylogenetic analysis. The optimized primer set for the 16S rRNA gene and the newly designed primer set for the hzo gene were successfully applied to identify anammox bacteria from marine sediments of aquaculture zone, coastal wetland, and deep ocean where the three ecosystems form a gradient of anthropogenic impact. Results indicated a broad distribution of anammox bacteria with high niche-specific community structure within each marine ecosystem.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Byrne N, Strous M, Crepeau V, Kartal B, Birrien JL, Schmid M, Lesongeur F, Schouten S, Jaeschke A, Jetten M, Prieur D, Godfroy A (2009) Presence and activity of anaerobic ammonium-oxidizing bacteria at deep-sea hydrothermal vents. ISME J 3:117–123
Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM (2005) The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33:D294–D296
Friedrich MW, Schmitt-Wagner D, Lueders T, Brune A (2001) Axial differences in community structure of Crenarchaeota and Euryarchaeota in the highly compartmentalized gut of the soil-feeding termite Cubitermes orthognathus. Appl Environ Microbiol 67:4880–4890
Klotz MG, Stein LY (2008) Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol Lett 278:146–156
Klotz MG, Schmid MC, Strous M, den Camp HJMO, Jetten MSM, Hooper AB (2008) Evolution of an octahaem cytochrome c protein family that is key to aerobic and anaerobic ammonia oxidation by bacteria. Environ Microbiol 10:3150–3163
Kuenen JG (2008) Anammox bacteria: from discovery to application. Nat Rev Microbiol 6:320–326
Lozupone C, Hamady M, Knight R (2006) UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371
Mulder A, van de Greef AA, Robertson LA, Kuenen JG (1995) Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol Ecol 16:177–184
Neef A, Amann R, Schlesner H, Schleifer KH (1998) Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257–3266
Penton CR, Devol AH, Tiedje JM (2006) Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl Environ Microbiol 72:6829–6832
Quan ZX, Rhee SK, Zuo JE, Yang Y, Bae JW, Park JR, Lee ST, Park YH (2008) Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environ Microbiol 10:3130–3139
Schalk J, de Vries S, Kuenen JG, Jetten MS (2000) Involvement of a novel hydroxylamine oxidoreductase in anaerobic ammonium oxidation. Biochemistry 39:5405–5412
Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, Metzger JW, Schleifer KH, Wagner M (2000) Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol 23:93–106
Schmid M, Walsh K, Webb R, Rijpstra WI, van de Pas-Schoonen K, Verbruggen MJ, Hill T, Moffett B, Fuerst J, Schouten S, Damste JS, Harris J, Shaw P, Jetten M, Strous M (2003) Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 26:529–538
Schmid MC, Maas B, Dapena A, van de Pas-Schoonen K, van de Vossenberg J, Kartal B, van Niftrik L, Schmidt I, Cirpus I, Kuenen JG, Wagner M, Sinninghe Damste JS, Kuypers M, Revsbech NP, Mendez R, Jetten MS, Strous M (2005) Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl Environ Microbiol 71:1677–1684
Schmid MC, Risgaard-Petersen N, van de Vossenberg J, Kuypers MM, Lavik G, Petersen J, Hulth S, Thamdrup B, Canfield D, Dalsgaard T, Rysgaard S, Sejr MK, Strous M, den Camp HJ, Jetten MS (2007) Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ Microbiol 9:1476–1484
Schmid MC, Hooper AB, Klotz MG, Woebken D, Lam P, Kuypers MM, Pommerening-Roeser A, Op den Camp HJ, Jetten MS (2008) Environmental detection of octahaem cytochrome c hydroxylamine/hydrazine oxidoreductase genes of aerobic and anaerobic ammonium-oxidizing bacteria. Environ Microbiol 10:3140–3149
Shimamura M, Nishiyama T, Shigetomo H, Toyomoto T, Kawahara Y, Furukawa K, Fujii T (2007) Isolation of a multiheme protein with features of a hydrazine-oxidizing enzyme from an anaerobic ammonium-oxidizing enrichment culture. Appl Environ Microbiol 73:1065–1072
Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Medigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJ, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes HW, Weissenbach J, Jetten MS, Wagner M, Le Paslier D (2006) Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440:790–794
Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MSM (1999) Missing lithotroph identified as new planctomycete. Nature 400:446–449
Tal Y, Watts JE, Schreier HJ (2005) Anaerobic ammonia-oxidizing bacteria and related activity in Baltimore inner harbor sediment. Appl Environ Microbiol 71:1816–1821
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
van de Graaf AA, Mulder A, de Bruijn P, Jetten MS, Robertson LA, Kuenen JG (1995) Anaerobic oxidation of ammonium is a biologically mediated process. Appl Environ Microbiol 61:1246–1251
Woebken D, Lam P, Kuypers MM, Naqvi SW, Kartal B, Strous M, Jetten MS, Fuchs BM, Amann R (2008) A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environ Microbiol 10:3106–3119
Acknowledgments
This research was supported by a Ph.D. studentship from The University of Hong Kong and in part by HKU765507M to J-DG and by the National Natural Science Foundation of China (3080032) and Guangdong Province Natural Science Foundation (84510301001692) to Y-GH. MGK was funded by incentive funds provided by the UofL-EVPR office and the US National Science Foundation (EF-0412129). We thank Jessie Lai for the laboratory assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Supplementary materials
(DOC 151 kb)
Rights and permissions
About this article
Cite this article
Li, M., Hong, Y., Klotz, M.G. et al. A comparison of primer sets for detecting 16S rRNA and hydrazine oxidoreductase genes of anaerobic ammonium-oxidizing bacteria in marine sediments. Appl Microbiol Biotechnol 86, 781–790 (2010). https://doi.org/10.1007/s00253-009-2361-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2361-5