Abstract
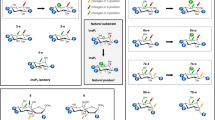
Inositol polyphosphatases (IPPases), particularly those that can hydrolyze myo-inositol hexakisphosphate (Ins P6), are of biotechnological interest for their ability to reduce the metabolically unavailable organic phosphate content of feedstuffs and to produce lower inositol polyphosphates (IPPs) for research and pharmaceutical applications. Here, the gene coding for a new protein tyrosine phosphatase (PTP)-like IPPase was cloned from Megasphaera elsdenii (phyAme), and the biochemical properties of the recombinant protein were determined. The deduced amino acid sequence of PhyAme is similar to known PTP-like IPPases (29–44% identity), and the recombinant enzyme displayed strict specificity for IPP substrates. Optimal IPPase activity was displayed at an ionic strength of 250 mM, a pH of 5.0, and a temperature of 60°C. In order to elucidate its stereospecificity of Ins P6 dephosphorylation, a combination of high-performance ion-pair chromatography and kinetic studies was conducted. PhyAme displayed a stereospecificity that is unique among enzymes belonging to this class in that it preferentially cleaved Ins P6 at one of two phosphate positions, 1D-3 or 1D-4. PhyAme followed two distinct and specific routes of hydrolysis, predominantly degrading Ins P6 to Ins(2)P via: (a) 1D-Ins(1,2,4,5,6)P5, 1D-Ins(1,2,5,6)P4, 1D-Ins(1,2,6)P3, and 1D-Ins(1,2)P2 (60%) and (b) 1D-Ins(1,2,3,5,6)P5, 1D-Ins(1,2,3,6)P4, Ins(1,2,3)P3, and d/l-Ins(1,2)P2 (35%).




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Baldi P, Pollastri G (2003) The principled design of large-scale recursive neural network architectures-dag-rnns and the protein structure prediction problem. J Mach Learn Res 4:575–603
Batten GD, Lott JNA (1986) The influence of phosphorus nutrition on the appearance and composition of globoid crystals in wheat aleurone cells. Cereal Chem 63:14–18
Bendtsen JD, Nielson H, Von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Billington DC (1993) The inositol phosphates. Chemical synthesis and biological significance. VCH, New York, NY
Brailoiu E, Miyamoto MD, Dun NJ (2003) Inositol derivatives modulate spontaneous transmitter release at the frog neuromuscular junction. Neuropharmacology 45:691–701
Brancaccio M, Legendre GG (1979) Megasphaera elsdenii Endocarditis. J Clin Microbiol 10:72–74
Cheng J, Randall AZ, Sweredoski MJ, Baldi P (2005) Scratch: a protein structure and structural feature prediction server. Nucl Acids Res 33:W72–W76
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the clustal series of programs. Nucl Acids Res 31:3497–3500
Cho CY, Koo SH, Wang Y, Callaway S, Hedrick S, Mak PA, Orth AP, Peters EC, Saez E, Montminy M, Schultz PG, Chanda SK (2006) Identification of the tyrosine phosphatase ptp-meg2 as an antagonist of hepatic insulin signaling. Cell Metab 3:367–378
Chu HM, Guo RT, Lin TW, Chou CC, Shr HL, Lai HL, Tang TY, Cheng KJ, Selinger BL, Wang AHJ (2004) Structures of Selenomonas ruminantium phytase in complex with persulfated phytate: DSP-phytase fold and mechanism for sequential substrate hydrolysis. Structure 12:2015–2024
Elsden SR, Volcani BE et al (1956) Properties of a fatty acid forming organism isolated from the rumen of sheep. J Bacteriol 72:681–689
Fassler J, Nadel C, Richardson N, McEntyre J, Schuler G, McGinnis S, Pongor S (2000) NCBI website. from http://www.ncbi.nlm.nih.gov/
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the expasy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, New York, pp 571–607
Greer J (1981) Comparative model-building of the mammalian serine proteases. J Mol Biol 153:1027–1042
Greiner R, Konietzny U (1996) Construction of a bioreactor to produce special breakdown products of phytate. J Biotechnol 48:153–159
Greiner R, Alminger ML, Carlsson NG, Muzquiz M, Burbano C, Cuadrado C, Pedrosa MM, Goyoaga C (2002a) Pathway of dephosphorylation of myo-inositol hexakisphosphate by phytases of legume seeds. J Agric Food Chem 50:6865–6870
Greiner R, Farouk A, Alminger ML, Carlsson NG (2002b) The pathway of dephosphorylation of myo-inositol hexakisphosphate by phytate-degrading enzymes of different Bacillus spp. Can J Microbiol 48:986–994
Haikara A, Helander I (2006) Pectinatus, Megasphaera and Zymophilus. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes: a handbook on the biology of bacteria, volume 4. Springer Science & Business Media, New York, pp 965–981
Hanakahi LA, Bartlet-Jones M, Chappell C, Pappin D, West SC (2000) Binding of inositol phosphate to DNA-pk and stimulation of double-strand break repair. Cell 102:721–729
Konietzny U, Greiner R (2002) Molecular and catalytic properties of phytate-degrading enzymes (phytases). Int J Food Sci Technol 37:791–812
Koppolu A, Clements LD (2004) Ruminal waste stream as a source of industrial chemicals. Resour Conserv Recycl 41:215–226
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lassen SF, Breinholt J, Ostergaard PR, Brugger R, Bischoff A, Wyss M, Fuglsang CC (2001) Expression, gene cloning, and characterization of five novel phytases from four basidiomycete fungi: Peniophora lycii, Agrocybe Pediades, ceriporia sp., and Trametes pubescens. Appl Environ Microbiol 67:4701–4707
Mullaney EJ, Ullah AHJ (2003) The term phytase comprises several different classes of enzymes. Biochem Biophys Res Commun 312:179–184
Nakashima BA, McAllister TA, Sharma R, Selinger LB (2007) Diversity of phytases in the rumen. Microb Ecol 53:82–88
Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: Analysis and visualization of genetic variation. from http://www.psc.edu/biomed/genedoc
Ohkawa T, Ebisuno S, Kitagawa M, Morimoto S, Miyazaki Y, Yasukawa S (1984) Rice bran treatment for patients with hypercalciuric stones: experimental and clinical studies. J Urol 132:1140–1145
Orchiston EA, Bennett D, Leslie NR, Clarke RG, Winward L, Downes CP, Safrany ST (2004) Pten m-cbr3, a versatile and selective regulator of inositol 1,3,4,5,6-pentakisphosphate (ins(1,3,4,5,6)p-5)—evidence for Ins(1,3,4,5,6)p-5 as a proliferative signal. J Biol Chem 279:1116–1122
Phillippy BQ, Bland JM (1988) Gradient ion chromatography of inositol phosphates. Anal Biochem 175:162–166
Pollastri G, Przybylski D, Rost B, Baldi P (2002) Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins 47:228–235
Priefer U, Simon R, Pühler A (1984) Cloning with cosmids. In: Pühler A, Timmis KN (eds) Advanced molecular genetics. Springer, New York, pp 190–201
Puhl AA, Gruninger RJ, Greiner R, Janzen TW, Mosimann SC, Selinger LB (2007) Kinetic and structural analysis of a bacterial protein tyrosine phosphatase-like myo-inositol polyphosphatase. Protein Sci 16:1368–1378
Puhl AA, Greiner R, Selinger LB (2008a) Kinetics, substrate specificity, and stereospecificity of two new protein tyrosine phosphatase-like inositol polyphosphatases from Selenomonas lacticifex. Biochem Cell Biol 86:322–330
Puhl AA, Greiner R, Selinger LB (2008b) A protein tyrosine phosphatase-like inositol polyphosphatase from Selenomonas ruminantium subsp. lactilytica has specificity for the 5-phosphate of myo-inositol hexakisphosphate. Int J Biochem Cell Biol 40:2053–2064
Ruf JC, Ciavatti M, Gustafsson T, Renaud S (1994) In-vitro effect of d-myo-inositol 1,2,6-trisphosphate (PP-56) on aggregation of platelets from normal and diabetic rats—relationship to malondialdehyde release and phosphoinositide pathway. Can J Physiol Pharmacol 72:644–649
Sasakawa N, Sharif M, Hanley MR (1995) Metabolism and biological-activities of inositol pentakisphosphate and inositol hexakisphosphate. Biochem Pharmacol 50:137–146
Scott HW, Dehority BA (1965) Vitamin requirements of several cellulolytic bacteria. J Bacteriol 89:1169–1175
Shears SB (2001) Assessing the omnipotence of inositol hexakisphosphate. Cell Signal 13:151–158
Skoglund E, Carlsson NG, Sandberg AS (1998) High-performance chromatographic separation of inositol phosphate isomers on strong anion exchange columns. J Agric Food Chem 46:1877–1882
Wohlt JF-, Sniffen CJ, Hoover WH (1973) Measurement of protein solubility in common feedstuffs. J Dairy Sci 56:1052–1057
Yanke LJ, Bae HD, Selinger LB, Cheng K-J (1998) Survey of phytase activity in anaerobic rumen bacteria. Microbiol 144:1565–1573
Zhang Z, Song Y, Wang XL (2005) Inositol hexaphosphate-induced enhancement of natural killer cell activity correlates with suppression of colon carcinogenesis in rats. World J Gastroenterol 11:5044–5046
Acknowledgements
L. Brent Selinger receives funding from the Natural Sciences and Engineering Research Council (NSERC) and the Advanced Foods and Materials Network (AFMNet). Thanks to L. J. Yanke, Agriculture and Agri-Food Canada (Lethbridge, Alberta), for supplying the M. elsdenii cultures. Analysis of the isomers of the individual myo-inositol phosphate derivatives by N.-G. Carlsson, Chalmers University of Technology (Göteborg, Sweden), is also gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Puhl, A.A., Greiner, R. & Selinger, L.B. Stereospecificity of myo-inositol hexakisphosphate hydrolysis by a protein tyrosine phosphatase-like inositol polyphosphatase from Megasphaera elsdenii . Appl Microbiol Biotechnol 82, 95–103 (2009). https://doi.org/10.1007/s00253-008-1734-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1734-5