Abstract
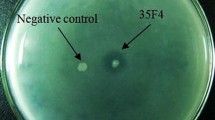
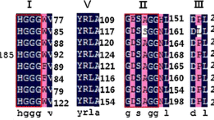
To search for new cold-active lipases, a metagenomic library was constructed using cold-sea sediment samples at Edison Seamount and was screened for lipolytic activities by plating on a tricaprylin medium. Subsequently, a fosmid clone was selected, and the whole sequence of 36 kb insert of the fosmid clone was determined by shotgun sequencing. The sequence analysis revealed the presence of 25 open reading frames (ORF), and ORF20 (EML1) showed similarities to lipases. Phylogenetic analysis of EML1 suggested that the protein belonged to a new family of esterase/lipase together with LipG. The EML1 gene was expressed in Escherichia coli, and purified by metal-chelating chromatography. The optimum activity of the purified EML1 (rEML1) occurred at pH 8.0 and 25°C, respectively, and rEML1 displayed more than 50% activity at 5°C. The activation energy for the hydrolysis of olive oil was determined to be 3.28 kcal/mol, indicating that EML1 is a cold-active lipase. rEML1 preferentially hydrolyzed triacylglycerols acyl-group chains with long chain lengths of ≥8 carbon atoms and displayed hydrolyzing activities toward various natural oil substrates. rEML1 was resistant to various detergents such as Triton X-100 and Tween 80. This study represents an example which developed a new cold-active lipase from a deep-sea sediment metagenome.




Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Arpigny JL, Jaeger KE (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343:177–183
Birnboim HC, Doly J (1979) A rapid alkaline procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523
Butman CA, Carlton JT (1995) Marine biological diversity-some important issues, opportunities and critical research needs. Rev Geophys 33:1201–1209
Choo DW, Tatsuo K, Takeshi S, Kenji S, Nobuyoshi E (1998) A cold-adapted lipase of an Alaskan Psychrotroph, Pseudomonas sp. strain B11–1: gene cloning and enzyme purification and characterization. Appl Environ Microbiol 64:486–491
Corliss JB, Dymond J, Gordon LI, Edmond JM, von Herzen RP, Ballard RD, Green K, Williams D, Bainbridge A, Crane K, van Andel TH (1979) Submarine thermal springs on the Galapagos Rift. Science 203:1073–1083
Duong F, Soscia C, Lazdundki A, Murgier M (1994) The Pseudomonas fluorescens lipase has a C-terminal secretion signal and is secreted by a three-component bacterial ABC-exporter system. Mol Microbiol 11:1117–1126
Elend C, Schmeisser C, Leggewie C, Babiak P, Carballeira JD, Steele HL, Reymond JL, Jaeger KE, Streit WR (2006) Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl Environ Microbiol 72:3637–3645
Feller G, Gerday C (2003) Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol 1:200–208
Feller G, Narinix E, Arpigny JL, Aittaleb M, Baise E, Genicot S, Gerday C (1996) Enzymes from psychrophilic organisms. FEMS Microbiol Rev 18:189–202
Gray JS (2002) Species richness of marine soft sediments. Mar Ecol Prog Ser 244:285–297
Gupta R, Beg QK, Lorenz P (2002) Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol 59:15–32
Handelsman J (2004) Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685
Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5:245–249
Herzig P, Hannington M, McInnes B, Stoffers P, Villinger H, Seifert R, Binns R, Liebe T (1994) Submarine volcanism and hydrothermal venting studied in Papua, New Guinea. EOS 75:513–520
Hurt RA, Qiu X, Wu L, Rou Y, Palumbo AV, Tiedje JM, Zhou J (2001) Simultaneous recovery of RNA and DNA from soils and sediments. Appl Environ Microbiol 67:4495–4503
Iwai M, Tsujisaka Y, Fukumoto J (1970) Studies on lipase. V. Effect of iron ions on the Aspergillus niger lipase. J Gen Appl Microbiol 16:81–90
Jaeger KE, Ransac S, Dijkstra BW, Colson C, van Heuvel M, Misset O (1994) Bacterial lipase. FEMS Microbiol Rev 15:29–63
Jaeger KE, Dijkstra BW, Reetz MT (1999) Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol 53:315–351
Kennicutt MC, Brooks JM, Bidigare RR, Fay RR, Wade TL, McDonald TJ (1985) Vent-type taxa in a hydrocarbon seep region on the Louisiana slope. Nature 317:351–353
Kim HK, Park SY, Lee JK, Oh TK (1998) Gene cloning and characterization of thermostable lipase from Bacillus stearothermophilus L1. Biosci Biotechnol Biochem 62:66–71
Kim JT, Kang SG, Woo JH, Lee JH, Jeong BC, Kim SJ (2007) Screening and its potential application of lipolytic activity from a marine environment: characterization of a novel esterase from Yarrowia lipolytica CL180. Appl Microbiol Biotechnol 74:820–828
Klibanov AM (2001) Improving enzymes by using them in organic solvents. Nature 409:241–246
Koeller KM, Wong CH (2001) Enzymes for chemical synthesis. Nature 409:232–240
Lee YP, Chung GH, Rhee JS (1993) Purification and characterization of Pseudomonas fluorescens SIK W1 lipase expressed in Escherichia coli. Biochim Biophys Acta 1169:156–164
Lee SH, Oh JR, Lee JH, Kim SJ, Cho JC (2004) Cold-seep sediment harbors phylogenetically diverse uncultured bacteria. J Microbiol Biotechnol 14:906–913
Lee MH, Lee CH, Oh TK, Song JK, Yoon JH (2006) Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl Environ Microbiol 72:7406–7409
Levin LA, Etter RJ, Rex MA, Gooday AJ, Smith CR, Pineda J, Stuart CT, Hessler RR, Pawson D (2001) Environmental influences on regional deep-sea species diversity. Annu Rev Ecol Syst 32:51–93
Liu W, Beppu T, Arima K (1973) Effect of various inhibitors on lipase action of thermophilic fungus Humicola lanuginosa S-38. Agric Biol Chem 37:2487–2492
Naeem R, Yuji S, Satoshi E, Haruyuki A, Tadayuki I (2001) Low-temperature lipase from Psychrotrophic Pseudomonas sp. strain KB700A. Appl Environ Microbiol 67:4064–4069
Ogino H, Nakagawa S, Shinya K, Muto T, Fujimura N, Yasudo M, Ishikawa H (2000) Purification and characterization of organic solvent tolerant lipase from organic solvent tolerant Pseudomonas aeruginosa LST-03. J Biosci Bioeng 89:451–457
Park SY, Kim JT, Kang SG, Woo JH, Lee JH, Choi HT, Kim SJ (2007) A new esterase showing similarity to putative dienelactone hydrolase from a strict marine bacterium, Vibrio sp. GMD509. Appl Microbiol Biotechnol 77:107–115
Rhee JK, Ahn DG, Kim YG, Oh JW (2005) New thermophilic and thermostable esterase with sequence similarity to the hormone-sensitive lipase family, cloned from a metagenomic library. Appl Environ Microbiol 71:817–825
Robertson DE, Chaplin JA, DeSantis G, Podar M, Madden M, Chi E, Richardson T, Milan A, Miller M, Weiner DP, Wong K, McQuaid J, Farwell B, Preston LA, Tan X, Snead MA, Keller M, Mathur E, Kretz PL, Burk MJ, Short JM (2004) Exploring nitrilase sequence space for enantioselective catalysis. Appl Environ Microbiol 70:2429–2436
Rosenau F, Jaeger KE (2000) Bacterial lipases from Pseudomonas: regulation of gene expression and mechanisms of secretion. Biochimie 82:1023–1032
Rosenberg M, Court D (1979) Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet 13:319–353
Ryu HS, Kim HK, Choi WC, Kim MH, Park SY, Han NS, Oh TK, Lee JK (2006) New cold-adapted lipase from Photobacterium lipolyticum sp. nov. that is closely related to filamentous fungal lipases. Appl Microbiol Biotechnol 70:321–326
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor
Schloss PD, Handelsman J (2003) Biotechnological prospects from metagenomics. Curr Opin Biotechnol 14:303–310
Schmidt RD, Verger R (1998) Lipases: interfacial enzymes with attractive applications. Angew Chem Int Ed Engl 37:1608–1633
Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B (2001) Industrial biocatalysis today and tomorrow. Nature 409:258–268
Solbak AI, Richardson TH, McCann RT, Kline KA, Bartnek F, Tomlinson G, Tan X, Parra-Gessert L, Frey GJ, Podar M, Luginbühl P, Gray KA, Mathur EJ, Robertson DE, Burk MJ, Hazlewood GP, Short JM, Kerovuo J (2005) Discovery of pectin-degrading enzymes and directed evolution of a novel pectate lyase for processing cotton fabric. J Biol Chem 280:9431–9438
Steele HL, Streit WR (2005) Metagenomics: advances in ecology and biotechnology. FEMS Microbiol Lett 247:105–111
Streit WR, Schmitz RA (2004) Metagenomics: the key to the uncultured microbes. Curr Opin Microbiol 7:492–498
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Villeneuve P, Muderhwa JM, Graille J, Haas MJ (2000) Customizing lipases for biocatalysis: a survey of chemical, physical and molecular biological approaches. J Mol Catal B Enzym 9:113–148
Voget S, Leggewie C, Uesbeck A, Raasch C, Jaeger KE, Streit WR (2003) Prospecting for novel biocatalysts in a soil metagenome. Appl Environ Microbiol 69:6235–6242
Yun J, Kang S, Park S, Yoon H, Kim MJ, Heu S, Ryu S (2004) Characterization of a novel amylolytic enzyme encoded by a gene from a soil-derived metagenomic library. Appl Environ Microbiol 70:7229–7235
Acknowledgement
This work was supported by KORDI in-house program (PE98230) and the Marine and Extreme Genome Research Center Program, Ministry of Marine Affairs and Fisheries, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jeon, J.H., Kim, JT., Kim, Y.J. et al. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl Microbiol Biotechnol 81, 865–874 (2009). https://doi.org/10.1007/s00253-008-1656-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1656-2