Abstract
The peptide repertoire presented on human leukocyte antigen (HLA) class I molecules is largely determined by the structure of the peptide binding groove. It is expected that the molecules having similar grooves (i.e., belonging to the same supertype) might present similar/overlapping peptides. However, the extent of promiscuity among HLA class I ligands remains controversial: while in many studies T cell responses are detected against epitopes presented by alternative molecules across HLA class I supertypes and loci, peptide elution studies report minute overlaps between the peptide repertoires of even related HLA molecules. To get more insight into the promiscuous peptide binding by HLA molecules, we analyzed the HLA peptide binding data from the large epitope repository, Immune Epitope Database (IEDB), and further performed in silico analysis to estimate the promiscuity at the population level. Both analyses suggest that an unexpectedly large fraction of HLA ligands (>50%) bind two or more HLA molecules, often across supertype or even loci. These results suggest that different HLA class I molecules can nevertheless present largely overlapping peptide sets, and that “functional” HLA polymorphism on individual and population level is probably much lower than previously anticipated.
Similar content being viewed by others
Introduction
The repertoire of the cytotoxic T cell response is shaped by the peptides presented on human leukocyte antigen (HLA) class I molecules. HLA class I genes are the most polymorphic loci known in the human genome: more than 2,000 HLA-A and -B allelic variants have been reported (Bjorkman and Parham 1990; Parham and Ohta 1996; Marsh et al. 2010). Most polymorphism is accumulated in the peptide binding groove of these molecules, giving rise to specific binding motifs for every HLA molecule, which allow for selective binding of a set of peptides forming the so called ligand-binding repertoire. HLA class I molecules may be grouped into several supertypes based on their potential binding motif similarity (Sette and Sidney 1999; Lund et al. 2004; Doytchinova et al. 2004; Kangueane et al. 2005; Reche and Reinherz 2007; Hertz and Yanover 2007; Sidney et al. 2008).
The specificity of HLA binding has been studied extensively in the last 30 years. It has become clear early on that some HLA molecules may significantly overlap in peptide binding specificity (Sidney et al. 1995; Barber et al. 1995; Doolan et al. 1997; Bertoni et al. 1997; Threlkeld et al. 1997), meaning that one peptide ligand has the ability to bind to several HLA molecules. The majority of ligand sharing was observed among molecules that have similar binding motifs and therefore would be assigned to the same HLA supertype, i.e., a form of promiscuity that may be considered as “expected” (Brusic et al. 2002; Ueno et al. 2002; Burrows et al. 2003; Sidney et al. 2003; Takedatsu et al. 2004; Frahm et al. 2005; Leslie et al. 2006). Few other reports showed promiscuity across supertypes or even loci; these findings were considered as “exceptions” (Sabbaj et al. 2003; Masemola et al. 2004). Recently, however, two systematic studies challenged this general view on promiscuity of HLA class I peptides and reported unexpected but also conflicting results. Frahm et al. (2007) tested T cell responses to 242 well-defined viral epitopes from HIV and EBV in 100 subjects and found that 95% of these epitopes elicited a T cell response in at least one individual not expressing the original restricting HLA molecule. The majority of potential alternative HLA molecules were not matched to the same HLA supertype or even the same locus as the original restricting HLA molecule. Shortly after this study, Hillen et al. (2008) reported only minute overlaps (3%) between the epitope repertoires of HLA molecules belonging to the B44 supertype, based on several hundreds of eluted peptides from nine members of the HLA-B44 supertype. This result was very surprising also because Sidney et al. (2003) reported largely overlapping peptide-binding repertoires for HLA molecules belonging to HLA-B44 family, based on in vitro MHC binding experiments. Of note, the experiments of Hillen et al. resulted in less than 30 peptides for some of the HLA molecules belonging to this supertype (e.g., B*5001, B*4701 and B*4501; see Hillen et al. 2008 and Table 3 of this article), suggesting that the peptide elution approach might underestimate the peptide binding repertoire of an HLA molecule.
Here we study the ligand sharing among HLA class I molecules by carrying out a systematic study, in which we analyze the data from Frahm et al. and Hillen et al. together with a large amount of data available from the IEDB (www.iedb.org; Vita et al. 2010) database. Although the experimental data in IEDB on MHC binding is extensive, it nevertheless does not provide a reliable estimate of promiscuous binding for every HLA molecule, because the number of HLA molecules that can bind the same peptide depends largely on the number of HLA molecules for which in vitro binding data is available for the peptide in question. To avoid this problem, we repeated the same analysis using state-of-the-art MHC class I binding predictors (Nielsen et al. 2007; Lundegaard et al. 2008; Hoof et al. 2009), where we estimate the extent of promiscuous peptide binding by taking into account every common HLA molecule in the population. In all cases, our results suggest that more than 60% of HLA ligands show promiscuous binding. Finally, we discuss consequences of the extensive ligand sharing among HLA class I molecules in the context of immunodominance and infectious diseases.
Results
HLA class I binding shows a high degree of promiscuity
To our knowledge, Frahm et al. (2007) were the first to study HLA class I binding promiscuity systematically. In short, a total of 242 known HIV-1 and EBV epitopes were tested in a cohort of 100 (50 HIV-1 infected and 50 healthy) subjects regardless of the individual's HLA type. This cohort had a diverse HLA distribution, covering 46 (common) HLA-A, -B, and -C molecules. Almost all of the tested epitopes, 95%, elicited a response in at least one individual not expressing the original restricting HLA molecule. Using two independent statistical approaches, Frahm et al. predicted the alternative HLA molecules. Surprisingly, the majority of potential alternative HLA molecules were outside the original restricting molecule’s supertype or even the locus (Frahm et al. 2007). Using the pan-specific MHC class I binding predictor NetMHCpan (Nielsen et al. 2007; Hoof et al. 2009), we confirmed 91% of these alternative HLA restrictions among the most significant associations and75% of all significant associations. This result suggests that the responses identified by Frahm et al. are largely due to promiscuous presentation of the same epitope via two or more HLA class I molecules, instead of possible T cell (CD4 and CD8) cross-reactivity to different (embedded) epitopes presented by HLA class I and II molecules. We observed that the predicted affinity for the alternative HLA molecules in the data of Frahm et al. is significantly lower than that for the original restricting HLA (p = 0.001, Mann–Whitney U test, for all associations). This may explain why the responses elicited by alternative HLA molecules could have been overseen so far, even though MHC-peptide binding at lower affinity does not necessarily result in lower T cell responses (Feltkamp et al. 1994; Sette et al. 1994; Fortier et al. 2008).
To test the HLA class I binding promiscuity in an independent data set, we analyzed HLA class I binding data from the IEDB database (Vita et al. 2010) (details are given in Materials and methods). This database covers approximately 99% of all publicly available information on peptide epitopes mapped in infectious agents. Obviously, the promiscuity of HLA binding depends on the number of different HLA alleles for which peptide binding is tested. To provide a realistic estimate of promiscuous HLA class I binding, we selected IEDB peptide epitopes for which in vitro binding assays were performed on at least six different HLA class I molecules. We will refer to this data set as "IEDB MHC binding data". With this criterion, a total of 3738 HLA class I binding peptides were retrieved, among which 72% were promiscuous, i.e., reported to bind to at least two HLA class I molecules (Table 1). Using a more stringent criterion, e.g., when including only the peptides which were tested on eight or ten HLA molecules, the average promiscuity remained high (>65%, results not shown). In line with the results of Frahm et al., 68% of promiscuous HLA class I binding was observed across serotypes, 47% across HLA supertypes, and 23% across HLA loci (Table 1). Although being a much smaller data set, CTL response data from IEDB suggests similar levels of promiscuity: Out of 135 non-redundant CTL epitopes, each of which was tested on at least six HLA alleles, 82 (60%) elicited responses in the context of two or more HLA molecules.
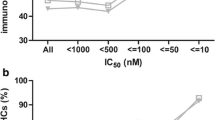
Finally, to estimate the promiscuous peptide binding on the population level, i.e., to estimate the chance of a peptide being presented by two or more HLA molecules in a population, we repeated a similar analysis using HLA binding predictors and focused on the most frequent 20 HLA-A and 20 HLA-B alleles in four US subpopulations with different ethnicity (European, Hispanic, African and Asian ethnicities, data extracted from the National Marrow Donor Program resource, http://bioinformatics.nmdp.org/; Maiers et al. 2007). We predicted potential 9-mer binders to these HLA molecules within common viral proteomes (n = 17, see Table S1) using NetMHCpan (Nielsen et al. 2007; Hoof et al. 2009). This prediction method was demonstrated to be the best one in a recent large benchmark performance test (Zhang et al. 2009). The most frequent HLA molecules (top 20 for A- and B-locus, respectively, listed in Table S4) in all four ethnic groups have, on average, a fraction of predicted promiscuous ligands around 60%, of which almost half are predicted to be presented by multiple HLA supertypes (Fig. 1). As expected, using a more stringent threshold to define the peptide binding (e.g., a predicted IC50 value of 50 nM instead of 500 nM) decreases the level promiscuity to 35–40%, as the ligand repertoire for each HLA molecule is severely reduced (results not shown). These results were reproducible with another neural network predictor, NetMHC3.2 (Lundegaard et al. 2008) (see Materials and Methods for a discussion on the choice of peptide–MHC binding predictors). Moreover, defining top 1–2% ranking binders as predicted binders changed only slightly the values reported in Table 1 and Fig. 1 (results not shown). In order to evaluate whether antigen processing has an impact on ligand promiscuity, we then added TAP and proteasomal cleavage predictions (Kesmir et al. 2002; Nielsen et al. 2005; Tenzer et al. 2005) to our MHC binding predictions. The level of promiscuity remained very similar (data not shown), implying that (predicted) antigen processing does not significantly influence the ligand sharing among HLA class I molecules. Taken together, not only the data reported by Frahm et al., but also HLA binding data available in the large IEDB repository and the analysis of HLA binding predictions on population level strongly suggest that a high fraction of HLA class I ligands (>60%) can bind to two or more HLA molecules and, frequently, the observed promiscuity occurs across different HLA class I supertypes.
Distribution of predicted HLA class I ligands of viral origin. All predicted ligands of the 20 most frequent HLA-A and HLA-B molecules in US subpopulations of a certain ethnic background (European, African, Asian and Hispanic) were classified into three categories: unique ligands (exclusively presented by one HLA class I molecule), within-supertype promiscuous ligands (exclusively targeted by one HLA supertype, but presented by at least two class I HLA molecules within this supertype) and across-supertype promiscuous ligands (targeted by HLA molecules belonging to at least two different HLA supertypes)
Next we detailed our analysis at the supertype level to pinpoint possible differences on ligand sharing among different HLA class I supertypes. Analyzing IEDB MHC binding data on per-supertype basis, we found that every supertype have exceptionally many peptides that exhibit promiscuous HLA binding (Table 2). The HLA-B44 supertype, analyzed by Hillen et al. (2008) is somewhat an “exception” among other supertypes: while 74% of the B44-ligands reported in IEDB exhibit promiscuous binding, across supertypes promiscuity is much lower at 17% (Table 2). A similar result was obtained in in silico analysis where we estimate the HLA peptide binding promiscuity on the population level (see Table S2).
Comparison of different experimental approaches for HLA peptide binding promiscuity
Hillen et al. (2008) undertook a different approach to study HLA class I binding promiscuity by directly comparing ligand repertoires based on peptides eluted from HLA molecules. As this is a very labor-intensive approach, they focused on a single supertype: HLA-B44, of which nine members were included in the elution study (listed in Table 3). Only a very small fraction (25 out of 670, 3%) of the “natural” ligands were found to bind two or more HLA molecules within the HLA-B44 supertype (Hillen et al. 2008). The binding promiscuity of ligands from different allele varies between 18% to none, with an average of 10% (Table 3). This unique data set allows us to compare the estimates of ligand sharing (for B44 supertype only) using three different experimental approaches: (i) T-cell binding (Frahm et al. 2007), (ii) eluted peptides (Hillen et al. 2008) and (iii) HLA–peptide binding measurements performed in vitro (IEDB data).
Frahm et al. tested the promiscuity of 20 CTL epitopes restricted by four HLA-B44 supertype members (Table 3). The far majority of those (17 epitopes) elicited a response in at least one individual negative for the original restricting allele. Five of these CTL epitopes elicited T cell responses in the context of another member of the HLA-B44 supertype (25% within-supertype promiscuity, Table 3). This result is similar to the one reported by Hillen et al., where out of four CD8+ T cell responses restricted by HLA-B44 molecules, only one epitope induced minor T cell responses in individuals negative for the restricting allele, but positive for a different HLA molecule within the B44 supertype (Hillen et al. 2008). The remaining 12 promiscuous B44 restricted CTL epitopes identified by Frahm et al. elicited responses in individuals that do not carry any HLA molecule belonging to the HLA-B44 supertype (60% outside-supertype promiscuity; Table 3).T cell response data extracted from IEDB show a similar trend: out of 14 T cell epitopes restricted by members of the B44 supertype and tested on at least six HLA molecules, only two (14%) show promiscuous binding within the supertype, while six epitopes (43%) elicit T cell responses when presented by an HLA molecule belonging to a different (non-B44) supertype.
Taken together, the results discussed above suggest that the estimates of HLA binding promiscuity for B44 supertype based on T cell responses (14–25% within supertype) are higher than the estimates based on eluted peptides (3% within supertype), and in vitro analysis of HLA binding provides the highest estimate of HLA binding promiscuity (see Table 2, 57% within supertype).
HLA-A and HLA-B molecules have similar levels of ligand promiscuity
So far we did not distinguish between different HLA loci in the promiscuity analysis. In the context of several infectious diseases, immune responses to epitopes restricted by HLA-B alleles were shown to be immunodominant (see, e.g., Kiepiela et al. 2004; Bihl et al. 2006). Moreover, particular HLA-B alleles seem to be associated with either protection or susceptibility to infectious diseases, best documented for HIV-1 infection (e.g., Carrington and O’Brien 2003; Kiepiela et al. 2004; Frahm et al. 2005; Pereyra et al. 2010). In order to see whether these features of HLA-B restricted T cell responses may be due to the promiscuous binding of HLA-B restricted epitopes, we compared their binding promiscuity to HLA-A restricted epitopes. Since Hillen et al. focused on HLA-B44 epitopes only, we used the data from Frahm et al. to perform this analysis. Frahm et al. tested 242 CTL epitopes tested, of which 148 and 181 epitopes were inferred to be presented by at least one HLA-A and -B molecule, respectively. The number of epitopes that were exclusively presented by a single HLA molecule was slightly higher for HLA-A than for HLA-B (HLA-A: 37 out of 148, HLA-B: 28 out of 181, p = 0.04, chi-square test), suggesting that HLA-B restricted epitopes might exhibit higher binding promiscuity. However, this difference between HLA-A and HLA-B epitopes was not to be found in IEDB T cell assay data (HLA-A: 45 out of 117, and HLA-B: eight out of 33, p = 0.19, chi-square test), and surprisingly, IEDB MHC binding data analysis suggested that HLA-A ligands have a higher level of binding promiscuity (p = 0.02, Mann–Whitney U test; Fig. S1).
Due to these conflicting results on the experimental data, we addressed the difference in the promiscuity of HLA-A and HLA-B ligands also by using HLA binding predictions. The fraction of peptides binding exclusively a single HLA molecule remained similar in predicted HLA-A and HLA-B ligands from viral proteomes (see Fig. S2, p = 0.34, Mann–Whitney U test). We repeated this analysis using a different set of criteria to define “binders” (i.e., using the top 1–2% percentile, see Materials and methods) and by extending our viral data set (Table S3), but in all cases we obtained similar results.
Taken together, since the available experimental data yield conflicting results and our in silico predictions suggest no significant difference in promiscuous peptide binding of HLA-A and HLA-B, we conclude that the ligand binding promiscuity probably does not play a major role in generating dominant HLA-B restricted responses.
Functional consequences of HLA peptide binding promiscuity in the context of HIV-1 infection
Our studies (see above) and others in the field provide solid evidence showing that HLA class I ligands show a high level of promiscuity. But why would HLA molecules have promiscuous ligand binding? After all, it is believed that the extensively polymorphic MHC has evolved due to a selective advantage of being able to present epitopes on rare MHC molecules in cases where the pathogens are (fully) adapted to common MHC molecules (Borghans et al. 2004). In search of a clue to explain functional aspects of such a high degree of promiscuity, we studied the effect of promiscuity on the disease outcome using HIV-1as a case study. We speculated that the individuals carrying HLA molecules with largely overlapping repertoires can be considered “functionally homozygous” and may therefore progress more rapidly to AIDS (Jeffery et al. 2000; Carrington and O’Brien 2003). We calculated the fraction of uniquely presented HIV-1 peptides for the frequent HLA alleles (based on the predicted peptide repertoires of the top 20 HLA-A and HLA-B molecules) in the Caucasian population. In line with the studies demonstrating that HLA-B alleles show the strongest association with disease outcome in HIV-1 infection (Kiepiela et al. 2004, 2007; Leslie et al. 2010),we found a strong negative correlation between the fraction of uniquely presented epitopes of HLA-B molecules and median viral loads reported by Fellay et al. (2009) (Fig. 2a; r = −0.57, p = 0.02, Spearman correlation test) or the relative hazard (RH) reported by Gao et al. (2001) (Fig. 2b; r = −0.60, p = 0.02, Spearman correlation test).Remarkably, some protective alleles, like B*2705 and B*5701, which have low RH and areassociated with low viral load, have more unique ligand repertoires than other alleles (82% and 56%, respectively, see Table S5), implying that having less promiscuous peptide presentation may contribute to viral control. However, when we repeated this analysis with data from the Durban cohort, infected mostly with HIV-1 clade C (Leslie et al. 2010), the fraction of uniquely presented epitopes no longer significantly correlated with the median viral loads (results not shown).
Correlation between the predicted fraction of unique ligands for HLA-B molecules and mean set point viral load associated with the same molecule or the relative hazard (RH).The fraction of uniquely presented HIV-1 peptides for an allele was calculated by comparing the predicted HIV-1 peptides for a particular allele with all the predicted HIV-1 peptides for the most frequent HLA alleles (top 20 HLA-A and HLA-B as listed in Table S4) in the Caucasian population. The predictions were performed with NetMHCpan (Hoof et al. 2009) to obtain data for as many as possible alleles. The correlation between the fraction of unique ligands and a mean set point viral load per HLA molecule taken from Fellay et al. (2009) and b RH taken from Gao et al. (2001) are shown. For (b), whenever available we have used allele specific RH; in other cases, we used the fraction of HIV-1 peptides estimated for the most dominant HLA-B allele to correlate with the relative hazard assigned to two digit HLA-B identifier (e.g., the relative hazard associated with B*40 is correlated with the fraction of unique HIV-1 peptides presented by HLA-B*4001). The Spearman correlation coefficients and corresponding significance values are reported in each figure. The data used to generate these graphs are given in Table S5
Discussion
The genes of the human major histocompatibilitycomplexbelong to the most polymorphic loci in the human population. However, it is not yet clear whether this large diversity at genotype level is reflected at the phenotype level by distinct ligand repertoires. It has been known for a long time that some HLA molecules have very similar binding motifs, and thus these molecules can be grouped into HLA supertypes (Sette and Sidney 1999; Lund et al. 2004; Doytchinova et al. 2004; Kangueane et al. 2005; Reche and Reinherz 2007; Hertz and Yanover 2007; Sidney et al. 2008). More recently, Frahm et al. demonstrated that promiscuous HLA class I binding reaches beyond the supertype: the far majority of HLA pairs that can elicit T cell responses to the same peptide belong to different supertypes. Following Frahm’s study on HIV-1 and EBV epitopes, it was demonstrated that human papillomavirus (HPV) and Mycobacterium tuberculosis (TB) epitopes also show extensive promiscuity of HLA class I binding when tested systematically (Nakagawa et al. 2007; Axelsson-Robertson et al. 2010). These findings were challenged by Hillen et al. (2008), who showed that even within a supertype, eluted HLA ligands can show as little as 3%promiscuity.
We have taken another approach to estimate HLA peptide binding promiscuity by using in vitro binding measurements reported in the IEDB database (Vita et al. 2010). Moreover, in order to be able to estimate the promiscuity of binding at the population level, i.e., by testing peptide binding to all major HLA molecules in a population, we performed in silico HLA–peptide binding predictions. In both cases we found extensive promiscuity in HLA class I ligand binding: 72% in the IEDB HLA binding data and 60% in our predicted HLA ligands. In addition, a high fraction of promiscuous ligands are found to be ligands for at least two different HLA supertypes (see Table 1, Fig. 1). As expected HLA supertype pairs with similar binding motifs share more ligands (e.g., A2 and B62 supertype ligands in IEDB overlap by 21%), than the pairs with dissimilar motifs (e.g., only 4.4% B27 supertype ligands in IEDB are also reported as binders for at least one allele belonging to B44 supertype).Since our in silico analysis covers different viruses and HLA molecules common in different ethnic groups, we believe that our results provide solid evidence for a high level of promiscuity being an intrinsic characteristic of HLA binding, regardless of the source of the ligand and the HLA molecule.
Why then was the fraction of shared ligands in the study by Hillen et al. as low as 3%? Unfortunately, our efforts did not produce a concrete answer to this question. Remarkably, the number of peptides eluted per allele by Hillen et al. was low (summarized in Table 2), considering that around 100,000 MHC molecules are expected to be on the cell surface at a time (Yewdell et al. 2003). This might be (among others) due to degradation of presented peptides under the rather harsh conditions necessary for the elution. If the elution studies underestimate the peptide repertoires, then the overlaps between the peptide repertoires of different MHC molecules might be underestimated as well. Indeed, when we use a stringent threshold to define binders in our in silico analysis, the predicted peptide repertoire of individual HLA molecules is reduced and as a consequence the average promiscuity decreases (data not shown).
By sampling eluted peptides, Hillen et al. may have biased their data to high affinity MHC binders. When we predicted the MHC binding affinity for the eluted peptides from the B44 supertype, we found that the median predicted binding affinity was lower than 50nM, which is generally used as a cutoff to discriminate high binders (e.g., for B*1801 the median affinity is 9 nM, and for B*4001 the median affinity is 27 nM). Following this, promiscuity of high affinity binders may be lower than of MHC ligands in general. However, this explanation is not in line with the earlier studies, which suggest that high affinity binders also tend to be the most promiscuous binders (Sidney et al. 1996, 2001; Sette et al. 2003). Similarly, we found a significant (but weak) negative correlation between ligand binding affinity and promiscuity (r = −0.27, p < 0.0001, Spearman correlation test), suggesting that the promiscuous binding among high affinity binders should be even higher than on average. Taken together, earlier studies and our present study suggest that the lower promiscuity observed by Hillen et al. might be due to other mechanisms than MHC binding per se.
The functional consequences of the extensive ligand sharing among HLA class I molecules remain to be discovered. In order to see whether promiscuous ligand presentation might be the underlying reason of immunodominance by HLA-B restricted T cell responses, we compared promiscuity between different HLA-A and HLA-B ligands. However, our numerous attempts did not result in a consistent picture (see section on HLA-A and -B), suggesting that there is not a direct association between (non-) promiscuous ligand presentation and dominant T cell responses. On the other hand, we have found a relationship between the fraction of uniquely presented peptides and HIV-1 disease progression, where HLA molecules associated with slow disease progression are also the ones that have the lowest degree of promiscuity (see Fig. 2). We believe that carrying HLA molecules with unique peptide repertoires increase the heterozygous advantage, based on the principle that individuals heterozygous at HLA loci are able to present a greater diversity of antigenic peptides than are homozygotes (Dean et al. 2002). The heterozygous advantage was suggested to generate a more effective immune response and therefore resulted in better control of HIV-1 infection (Carrington et al. 1999). In addition, the individuals with HLA molecules having unique binding motifs have lower chances of transmission of a pre-adapted virus. More data on HLA associations and disease outcome will help to resolve the functional aspects of the high level of promiscuity among HLA class I epitopes and especially how it affects an individual’s fitness.
Materials and methods
Experimental MHC binding and T cell response data
The experimental data used in our analysis was extracted from the Immune Epitope Database and Analysis Resource (IEDB; www.immuneepitope.org; downloads were made in March 2010). The first data set included all peptides for which the HLA class I binding affinity was determined by in vitro MHC binding assays; the second data set consisted of peptides with measured T cell responses. We considered only peptidesthatweretested on at least six HLA class I molecules and with an IC50 value lower than 500 nM for at least one of these molecules (i.e., the peptide has to be a binder for at least one HLA molecule). In addition, to make sure that all HLA–peptide associations were well defined, only the data with four-digit HLA class I identifierswere included. These selection criteria resulted in a set of 3,738 non-redundant peptides obtained from the MHC binding assay database of IEDB and is herereferred to as “IEDB MHC binding data”. The T cell response data, filtered using the same criteria, resulted in a much smaller data set. Therefore, we relaxed the requirements on four digit HLA identifiers by including T cell response data for which only one- or two-digit HLA identifiers were available. In total, filtering of the IEDB T cell assay data resulted in 135 non-redundant T cell epitopes.
Quantifying HLA binding promiscuity
We divided the ligands of a particular HLA class I molecule into two groups: (i) unique ligands, which are exclusively presented by this HLA class I molecule; (ii) promiscuous ligands, which are capable to bind to at least one other HLA class I molecule. We define the fraction of the unique ligands as, \( {F_{\text{u}}} = \frac{{{N_{\text{u}}}}}{{{N_{\text{all}}}}} \), where N u is the number of unique ligands and N all is the total number of ligands. In order to estimate this fraction as reliably as possible, we calculated it only for the HLA molecules for which more than ten peptides were experimentally tested on alternative HLA molecules. Changing this arbitrary threshold of 10 (to 20 or 30) did not change the results reported in the text (data not shown).
Promiscuity at supertype level
Throughout our analysis, we followed allele-supertype associations defined by Sidney et al. (2008) except in a single case: HLA-B*4901 is not classified into any supertype by Sidney et al., and therefore, we assigned it to the B44 supertype, as was done by Hillen et al. (2008).
In silico analysis
HLA allele selection
HLA allele frequencies were obtained from the National Marrow Donor Program (NMDP) website (bioinformatics.nmdp.org) for four predominant US census categories of race and ethnicity: African Americans, Asians, European Americans and Hispanics (Mori et al. 1997; Maiers et al. 2007). We included the 20 most frequent HLA-A and 20 most frequent HLA-B alleles for each ethnic group into our in silico analysis (Table S4A, S4B). The peptide–MHC binding predictions for majority of these molecules are of high quality (Hoof et al. 2009).
HLA class I ligand prediction
To have an as large as possible population coverage, we used NetMHCpan (Hoof et al. 2009) to predict peptide–HLA binding affinity. NetMHCpan assigns to each peptide–HLA pair a predicted IC50 value, indicative of the predicted binding affinity. An IC50 threshold of 500 nM was used to discriminate HLA binding ligands from nonbinding peptide. NetMHCpan is not an allele specific method: it has been trained on peptide binding data for many different MHC molecules (also from non-human species), and its prediction relies on intra- and extrapolation from characterized to uncharacterized HLA alleles. Thus, NetMHCpan may overestimate the promiscuity of HLA class I peptides. In order to check this issue, we compared NetMHCpan with an allele-specific predictor NetMHC3.2 (Buus et al. 2003; Nielsen et al. 2008; Lundegaard et al. 2008). A total of 42 HLA molecules (21 HLA-A and 21 HLA-B) have NetMHC3.2 predictions available. For each HLA molecule, we calculated the fraction of unique ligands to estimate the promiscuity of its ligands, by using the prediction results obtained from NetMHC3.2 and NetMHCpan. Predicted promiscuity of HLA class I binding by both predictors is highly correlated (p < 0.001, r = 0.86, Pearson correlation test). Moreover, both predictors estimate the fraction of promiscuous ligands restricted by these 42 HLA alleles to be around 58–60% (data not shown and Fig. 1). These results suggest that using NetMHCpan, which has a broader population coverage than NetMHC, would not result in an overestimation of promiscuity of HLA ligands.
Using a fixed threshold of 500 nM IC50 to define predicted binders may result in differences in predicted repertoire sizes between HLA molecules, which in turn may introduce a bias into the promiscuity analysis (MacNamara et al. 2009). To avoid this, we repeated the analysis by defining the top 1% of the peptides as candidate binders for each HLA molecule, thereby ensuring the same ligand repertoire size for each HLA molecule. With this alternative scaled threshold approach, all in silico results reported in this paper remain unchanged. For example, for the European subpopulation the predicted promiscuity of HLA class I binders is 57% with the fixed threshold of 500 nM and 54% with the scaled threshold.
Predicting antigen processing
The Stabilized Matrix Method (SMM) was applied to predict TAP transport efficiency and proteasomal cleavage, which are the two main steps of antigen processing (Tenzer et al. 2005). Applying an alternative predictor of antigen processing, NetChop (Kesmir et al. 2002; Nielsen et al. 2005), did not affect our results.
Viral data
The proteomes of 17 common human viruses were downloaded from the European Bioinformatics Institute website (www.ebi.ac.uk; downloads were made in Oct 2006, listed in Table S1) as the source of potential HLA ligands. We used the HLA, TAP and proteasome predictors to screen all possible unique virus-derived 9-mer peptides for potential HLA ligands. This data set is later extended with the viruses given in Table S3 to test the dependence of our results on the initial set of viral proteomes.
References
Axelsson-Robertson R, Weichold F, Sizemore D, Wulf M, Skeiky YAW, Sadoff J, Maeurer MJ (2010) Extensive major histocompatibility complex class I binding promiscuity for Mycobacterium tuberculosis TB10.4 peptides and immune dominance of human leucocyte antigen (HLA)-B*0702 and HLA-B*0801 alleles in TB10.4 CD8 T-cell responses. Immunology 129:496–505
Barber LD, Gillece-Castro B, Percival L, Li X, Clayberger C, Parham P (1995) Overlap in the repertoires of peptides bound in vivo by a group of related class I HLA-B allotypes. Curr Biol 5:179–190
Bertoni R, Sidney J, Fowler P, Chesnut RW, Chisari FV, Sette A (1997) Human histocompatibility leukocyte antigen-binding supermotifs predict broadly cross-reactive cytotoxic T lymphocyte responses in patients with acute hepatitis. J Clin Invest 100:503–513
Bihl F, Frahm N, Di Giammarino L, Sidney J, John M, Yusim K, Woodberry T, Sango K, Hewitt HS, Henry L et al (2006) Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol 176:4094–4101
Bjorkman PJ, Parham P (1990) Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem 59:253–288
Borghans JAM, Beltman JB, De Boer RJ (2004) MHC polymorphism under host–pathogen coevolution. Immunogenetics 55:732–739
Brusic V, Petrovsky N, Zhang G, Bajic VB (2002) Prediction of promiscuous peptides that bind HLA class I molecules. Immunol Cell Biol 80:280–285
Burrows SR, Elkington RA, Miles JJ, Green KJ, Walker S, Haryana SM, Moss DJ, Dunckley H, Burrows JM, Khanna R (2003) Promiscuous CTL recognition of viral epitopes on multiple human leukocyte antigens: biological validation of the proposed HLA A24 supertype. J Immunol 171:1407–1412
Buus S, Lauemoller SL, Worning P, Kesmir C, Frimurer T, Corbet S, Fomsgaard A, Hilden J, Holm A, Brunak S (2003) Sensitive quantitative predictions of peptide–MHC binding by a “Query by Committee” artificial neural network approach. Tissue Antigens 62:378–384
Carrington M, O’Brien SJ (2003) The influence of HLA genotype on AIDS. Annu Rev Med 54:535–551
Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O’Brien SJ (1999) HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748–1752
Dean M, Carrington M, O’Brien SJ (2002) Balanced polymorphism selected by genetic versus infectious human disease. Annu Rev Genomics Hum Genet 3:263–292
Doolan DL, Hoffman SL, Southwood S, Wentworth PA, Sidney J, Chesnut RW, Keogh E, Appella E, Nutman TB, Lal AA et al (1997) Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity 7:97–112
Doytchinova IA, Guan P, Flower DR (2004) Identifiying human MHC supertypes using bioinformatic methods. J Immunol 172:4314–4323
Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, Urban TJ, Zhang K, Gumbs CE, Smith JP et al (2009) Common genetic variation and the control of HIV-1 in humans. PLoS Genet 5:e1000791
Feltkamp MC, Vierboom MP, Kast WM, Melief CJ (1994) Efficient MHC class I-peptide binding is required but does not ensure MHC class I-restricted immunogenicity. Mol Immunol 31:1391–1401
Fortier M-H, Caron E, Hardy M-P, Voisin G, Lemieux S, Perreault C, Thibault P (2008) The MHC class I peptide repertoire is molded by the transcriptome. J Exp Med 205:595–610
Frahm N, Adams S, Kiepiela P, Linde CH, Hewitt HS, Lichterfeld M, Sango K, Brown NV, Pae E, Wurcel AG et al (2005) HLA-B63 presents HLA-B57/B58-restricted cytotoxic T-lymphocyte epitopes and is associated with low human immunodeficiency virus load. J Virol 79:10218–10225
Frahm N, Yusim K, Suscovich TJ, Adams S, Sidney J, Hraber P, Hewitt HS, Linde CH, Kavanagh DG, Woodberry T et al (2007) Extensive HLA class I allele promiscuity among viral CTL epitopes. Eur J Immunol 37:2419–2433
Gao X, Nelson GW, Karacki P, Martin MP, Phair J, Kaslow R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D et al (2001) Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med 344:1668–1675
Hertz T, Yanover C (2007) Identifying HLA supertypes by learning distance functions. Bioinformatics 23:e148–e155
Hillen N, Mester G, Lemmel C, Weinzierl AO, Müller M, Wernet D, Hennenlotter J, Stenzl A, Rammensee H-G, Stevanović S (2008) Essential differences in ligand presentation and T cell epitope recognition among HLA molecules of the HLA-B44 supertype. Eur J Immunol 38:2993–3003
Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M (2009) NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61:1–13
Jeffery KJ, Siddiqui AA, Bunce M, Lloyd AL, Vine AM, Witkover AD, Izumo S, Usuku K, Welsh KI, Osame M et al (2000) The influence of HLA class I alleles and heterozygosity on the outcome of human T cell lymphotropic virus type I infection. J Immunol 165:7278–7284
Kangueane P, Sakharkar MK, Rajaseger G, Bolisetty S, Sivasekari B, Zhao B, Ravichandran M, Shapshak P, Subbiah S (2005) A framework to sub-type HLA supertypes. Front Biosci 10:879–886
Kesmir C, Nussbaum AK, Schild H, Detours V, Brunak S (2002) Prediction of proteasome cleavage motifs by neural networks. Protein Eng 15:287–296
Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L et al (2004) Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775
Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, De Pierres C, Mncube Z, Mkhwanazi N et al (2007) CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 13:46–53
Leslie A, Price DA, Mkhize P, Bishop K, Rathod A, Day C, Crawford H, Honeyborne I, Asher TE, Luzzi G et al (2006) Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J Immunol 177:4699–4708
Leslie A, Matthews PC, Listgarten J, Carlson JM, Kadie C, Ndung’u T, Brander C, Coovadia H, Walker BD, Heckerman D et al (2010) Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol 84:9879–9888
Lund O, Nielsen M, Kesmir C, Petersen AG, Lundegaard C, Worning P, Sylvester-Hvid C, Lamberth K, Roder G, Justesen S et al (2004) Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics 55:797–810
Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M (2008) NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res 36:W509–W512
MacNamara A, Kadolsky U, Bangham CR, Asquith B (2009) T-cell epitope prediction: rescaling can mask biological variation between MHC molecules. PLoS Comput Biol 5:e1000327
Maiers M, Gragert L, Klitz W (2007) High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol 68:779–788
Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernández-Viña M, Geraghty DE, Holdsworth R, Hurley CK et al (2010) Nomenclature for factors of the HLA system, 2010. Tissue Antigens 75:291–455
Masemola AM, Mashishi TN, Khoury G, Bredell H, Paximadis M, Mathebula T, Barkhan D, Puren A, Vardas E, Colvin M et al (2004) Novel and promiscuous CTL epitopes in conserved regions of Gag targeted by individuals with early subtype C HIV type 1 infection from southern Africa. J Immunol 173:4607–4617
Mori M, Beatty PG, Graves M, Boucher KM, Milford EL (1997) HLA gene and haplotype frequencies in the North American population: the National Marrow Donor Program Donor Registry. Transplantation 64:1017–1027
Nakagawa M, Kim KH, Gillam TM, Moscicki A-B (2007) HLA class I binding promiscuity of the CD8 T-cell epitopes of human papillomavirus type 16 E6 protein. J Virol 81:1412–1423
Nielsen M, Lundegaard C, Lund O, Kesmir C (2005) The role of the proteasome in generating cytotoxic T cell epitopes: Insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57:33–41
Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, Roder G, Peters B, Sette A, Lund O et al (2007) NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS ONE 2:e796
Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S, Buus S, Lund O (2008) Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol 4:e1000107
Parham P, Ohta T (1996) Population biology of antigen presentation by MHC class I molecules. Science 272:67–74
Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PIW, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M et al (2010) The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557
Reche PA, Reinherz EL (2007) Definition of MHC supertypes through clustering of MHC peptide-binding repertoires. Methods Mol Biol 409:163–173
Sabbaj S, Bansal A, Ritter GD, Perkins C, Edwards BH, Gough E, Tang J, Szinger JJ, Korber B, Wilson CM et al (2003) Cross-reactive CD8+ T cell epitopes identified in US adolescent minorities. J Acquir Immune Defic Syndr 33:426–438
Sette A, Sidney J (1999) Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50:201–212
Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast WM, Melief CJ, Oseroff C, Yuan L, Ruppert J et al (1994) The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol 153:5586–5592
Sette A, Sidney J, Livingston BD, Dzuris JL, Crimi C, Walker CM, Southwood S, Collins EJ, Hughes AL (2003) Class I molecules with similar peptide-binding specificities are the result of both common ancestry and convergent evolution. Immunogenetics 54:830–841
Sidney J, del Guercio MF, Southwood S, Engelhard VH, Appella E, Rammensee HG, Falk K, Rötzschke O, Takiguchi M, Kubo RT (1995) Several HLA alleles share overlapping peptide specificities. J Immunol 154:247–259
Sidney J, Southwood S, del Guercio MF, Grey HM, Chesnut RW, Kubo RT, Sette A (1996) Specificity and degeneracy in peptide binding to HLA-B7-like class I molecules. J Immunol 157:3480–3490
Sidney J, Southwood S, Mann DL, Fernandez-Vina MA, Newman MJ, Sette A (2001) Majority of peptides binding HLA-A*0201 with high affinity crossreact with other A2-supertype molecules. Hum Immunol 62:1200–1216
Sidney J, Southwood S, Pasquetto V, Sette A (2003) Simultaneous prediction of binding capacity for multiple molecules of the HLA B44 supertype. J Immunol 171:5964–5974
Sidney J, Peters B, Frahm N, Brander C, Sette A (2008) HLA class I supertypes: a revised and updated classification. BMC Immunol 9:1
Takedatsu H, Shichijo S, Katagiri K, Sawamizu H, Sata M, Itoh K (2004) Identification of peptide vaccine candidates sharing among HLA-A3+, -A11+, -A31+, and -A33+ cancer patients. Clin Cancer Res 10:1112–1120
Tenzer S, Peters B, Bulik S, Schoor O, Lemmel C, Schatz MM, Kloetzel P-M, Rammensee H-G, Schild H, Holzhütter H-G (2005) Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol Life Sci 62:1025–1037
Threlkeld SC, Wentworth PA, Kalams SA, Wilkes BM, Ruhl DJ, Keogh E, Sidney J, Southwood S, Walker BD, Sette A (1997) Degenerate and promiscuous recognition by CTL of peptides presented by the MHC class I A3-like superfamily: implications for vaccine development. J Immunol 159:1648–1657
Ueno T, Tomiyama H, Takiguchi M (2002) Single T cell receptor-mediated recognition of an identical HIV-derived peptide presented by multiple HLA class I molecules. J Immunol 169:4961–4969
Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B (2010) The immune epitope database 2.0. Nucleic Acids Res 38:D854–D862
Yewdell JW, Reits E, Neefjes J (2003) Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immunol 3:952–961
Zhang H, Lundegaard C, Nielsen M (2009) Pan-specific MHC class I predictors: a benchmark of HLA class I pan-specific prediction methods. Bioinformatics 25:83–89
Acknowledgements
We thank Rob de Boer for valuable comments on the manuscript, Hanneke van Deutekom, Jorg Calis and Boris Schmid for technical support. This work was financially supported by a HIPO (High Potential) grant from Universiteit Utrecht.
Conflict of interest
The authors have no financial conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Promiscuity of HLA-A and HLA-B ligands based on IEDB MHC binding data. 30 HLA-A and 26 HLA-B molecules (each of which with at least 10 ligands in IEDB MHC binding data) were selected. The fraction of unique ligands was calculated for each allele. HLA-B ligands are significantly less promiscuous than HLA-A ligands (p = 0.02, Mann-Whitney U test). (DOC 37 kb)
Fig. S2
Promiscuity of predicted HLA-A and HLA-B ligands based on NetMHCpan predictions. All possible 9-mer viral ligands from viral proteomes (listed in Table S1 and S3) were predicted for the 20 most frequent HLA-A and HLA-B alleles in USA subpopulation of European ethnic background. The fraction of unique ligands (in percentages) was calculated for each allele (median of the group is indicated by a horizontal line). There is no significant difference between the fraction of unique HLA-A and HLA-B ligands (p = 0.34, Mann-Whitney U test). (DOC 38 kb)
Table S1
List of viral proteomes used as the resource of NetMHCpan predicted ligands. (DOC 37 kb)
Table S2
Ligand promiscuity of HLA supertypes for different ethnic groups based on NetMHCpan prediction data. We obtained very similar results with NetMHC3.2 predictor. (DOC 79 kb)
Table S3
List of human viral proteomes used in the extension of the viral set. (DOC 35 kb)
Table S4
A Most frequent 20 HLA-A alleles in four ethnic groups selected for NetMHCpan predictions. B Most frequent 20 HLA-B alleles in four ethnic groups selected for NetMHCpan predictions. (DOC 137 kb)
Table S5
Fraction of uniquely presented HIV-1 peptides by frequent HLA-B molecules, and the corresponding set point viral loads (mean) as reported by Fellay et al. 2009, and relative hazard (RH) values reported by Gao et al. 2001. The predictions reported here were made with NetMHCpan, however, similar results are obtained using allele specific predictor NetMHC3.2. Only the alleles where a reliable peptide binding predictor is available (defined as an estimated correlation coefficient between predicted and measured peptide binding affinities larger than 0.6) are included in the analysis. This data is used to make the analysis summarized in Fig. 2 of the main text. (DOC 39 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Rao, X., Hoof, I., Fontaine Costa, A.I.C.A. et al. HLA class I allele promiscuity revisited. Immunogenetics 63, 691–701 (2011). https://doi.org/10.1007/s00251-011-0552-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-011-0552-6