Abstract
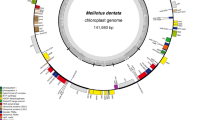
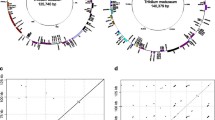
The plastid genome of Trifolium subterraneum is 144,763 bp, about 20 kb longer than those of closely related legumes, which also lost one copy of the large inverted repeat (IR). The genome has undergone extensive genomic reconfiguration, including the loss of six genes (accD, infA, rpl22, rps16, rps18, and ycf1) and two introns (clpP and rps12) and numerous gene order changes, attributable to 14–18 inversions. All endpoints of rearranged gene clusters are flanked by repeated sequences, tRNAs, or pseudogenes. One unusual feature of the Trifolium subterraneum genome is the large number of dispersed repeats, which comprise 19.5% (ca. 28 kb) of the genome (versus about 4% for other angiosperms) and account for part of the increase in genome size. Nine genes (psbT, rbcL, clpP, rps3, rpl23, atpB, psbN, trnI-cau, and ycf3) have also been duplicated either partially or completely. rpl23 is the most highly duplicated gene, with portions of this gene duplicated six times. Comparisons of the Trifolium plastid genome with the Plant Repeat Database and searches for flanking inverted repeats suggest that the high incidence of dispersed repeats and rearrangements is not likely the result of transposition. Trifolium has 19.5 kb of unique DNA distributed among 160 fragments ranging in size from 30 to 494 bp, greatly surpassing the other five sequenced legume plastid genomes in novel DNA content. At least some of this unique DNA may represent horizontal transfer from bacterial genomes. These unusual features provide direction for the development of more complex models of plastid genome evolution.






Similar content being viewed by others
References
Bookjans G, Stummann BM, Henningsen KW (1984) Preparation of chloroplast DNA from pea plastids isolated in a medium of high ionic strength. Anal Biochem 141:244–247
Bowman CM, Dyer TA (1986) The location and possible evolutionary significance of small dispersed repeats in wheat ctDNA. Curr Genet 10:931–941
Chang C-C, Lin H-C, Lin I-P, Chow T-Y, Chen H-H, Chen W-H, Cheng C-H, Lin C-Y, Liu S-M, Chang C-C, Chaw S-M (2006) The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol 23:279–291
Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK (2006) The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol 23:2175–2190
Cosner ME, Jansen RK, Palmer JD, Downie SR (1997) The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr Genet 31:419–429
Cosner ME, Raubeson LA, Jansen RK (2004) Chloroplast DNA rearrangements in Campanulaceae: phylogenetic utility of highly rearranged genomes. BMC Evol Biol 4:1–17
Darling ACE, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genet Res 14:1394–1403
Doyle JJ, Doyle JL, Palmer JD (1995) Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst Bot 20:272–294
Doyle JJ, Doyle JL, Palmer JD (1996) The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol Phylogenet Evol 5:429–438
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred II. error probabilities. Genome Res 8:186–194
Fan WH, Woelfle MA, Mosig G (1995) Two copies of a DNA element, Wendy, in the chloroplast chromosome of Chlamydomonas reinhardtii between rearranged gene clusters. Plant Mol Biol 29:63–80
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH (2003) Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol Biol Evol 20:1499–1505
Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH (2004) The chloroplast genome of Nymphaea alba: whole-genome analyses and the problem of identifying the most basal angiosperm. Mol Biol Evol 21:1445–1454
Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH (2005) Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol 22:1813–1822
Haberle RC, Fourcade HM, Boore JL, Jansen RK (2008) Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol 66:350–361
Hansen DR, Dastidar SG, Cai Z, Penaflor C, Kuehl JV, Boore JL, Jansen RK (2007) Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phylogenet Evol 45:547–563
Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D (2004) Type V protein secretion pathway: the autotransporter story. Micrbiol Mol Biol Rev 68:692–744
Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY, Li YQ, Kanno A, Nishizawa Y, Hirai A, Shinozaki K, Sugiura M (1989) The complete sequence of the rice Oryza sativa chloroplast genome - intermolecular recombination between distinct transfer-RNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet 217:185–194
Hupfer H, Swiatek M, Hornung S, Herrmann RG, Maier RM, Chiu WL, Sears B (2000) Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable Euoenothera plastomes. Mol Gen Genet 263:581–585
Jansen RK, Raubeson LA, Boore JL, dePamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson A, Peery R, Herman SJ, Fourcade HM, Kuehl JV, McNeal JR, Leebens-Mack J, Cui L (2005) Methods for obtaining and analyzing whole chloroplast genome sequences Molecular evolution: producing the biochemical data part B. Methods Enzymol 395:348–384
Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee S-B, Peery R, McNeal J, Kuehl JV, Boore JL (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA 104:19369–19374
Jansen RK, Wojciechowski MF, Sanniyasi E, Lee S-B, Daniell H (2008) Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Fabaceae). Mol Phylogenet Evol 48:1204–1217
Katayama H, Ogihara Y (1996) Phylogenetic affinities of the grasses to other monocots as revealed by molecular analysis of chloroplast DNA. Curr Genet 29:572–581
Knox EB, Palmer JD (1999) The chloroplast genome arrangement Lobelia thuliniana Lobeliaceae: expansion of the inverted repeat in an ancestor of the Campanulales. Plant Sys Evol 214:49–64
Lee H-L, Jansen RK, Chumley TW, Kim K-J (2007) Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol 24:1161–1180
Leebens-Mack J, Raubeson LA, Cui LY, Kuehl JV, Fourcade MH, Chumley TW, Boore JL, Jansen RK, dePamphilis CW (2005) Identifying the basal angiosperm node in chloroplast genome phylogenies: sampling one’s way out of the Felsenstein zone. Mol Biol Evol 22:1948–1963
McCarthy EM, McDonald JF (2003) LTR STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19:362–367
Meyers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11:1660–1676
Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, Kavanagh TA, Hibberd JM, Gray JC, Morden CW, Caile PJ, Jermiin LS, Wolfe KH (2001) Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plt Cell 13:645–658
Milligan BG, Hampton JN, Palmer JD (1989) Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol Biol Evol 6:355–368
Moore MJ, Dhingra A, Soltis P, Shaw R, Farmerie WG, Folta KM, Soltis DE (2006) Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plt Biol 6:17
Moore MJ, Bell CD, Soltis PS, Soltis DE (2007) Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA 104:19363–19368
Palmer JD (1986) Isolation and structural analysis of chloroplast DNA. Methods Enzymol 118:167–186
Palmer JD, Thompson WF (1981) Rearrangements in the chloroplast genomes of mung bean and pea. Proc Natl Acad Sci USA 78:5533–5537
Pombert J-F, Otis C, Lemieux C, Turmel M (2005) The chloroplast genome sequence of the green alga Pseudendoclonium `akinetum (Ulvophyceae) reveals unusual structural features and new insights into the branching order of chlorophyte lineages. Mol Biol Evol 22:1903–1918
Pombert J-F, Lemieux C, Turmel M (2006) The complete chloroplast DNA sequence of the green alga Oltmannsiellopsis viridis reveals a distinctive quadripartite architecture in the chloroplast genome of early diverging ulvophytes. BMC Biol 4:3
Raubeson LA, Peery R, Chumley T, Dziubek C, Fourcade HM, Boore JL, Jansen RK (2007) Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 8:174
Rice DW, Palmer JD (2006) An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Evol Biol 4:31
San Miguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot 82:37–44
Saski C, Lee S-B, Daniell H, Wood TC, Tomkins J, Kim H-G, Jansen RK (2005) Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol 59:309–322
Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchishinozaki K, Ohto C, Torazawa K, Y. Meng B, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M (1986) The complete nucleotide sequence of the tobacco chloroplast genome—its gene organization and expression. EMBO J 5:2043–2049
Tesler G (2002) GRIMM: genome rearrangements web server. Bioinformatics 18:492–493
Vicient CM, Suoniemi A, Anamthawat-Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH (1999) Retrotransposon BARE–1 and its role in genome evolution in the genus Hordeum. Plt Cell 11:1769–1784
Wang R-J, Cheng C-L, Chang C-C, Wu C-L, Su T-M, Chaw SM (2008) Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol 8:36
Wojciechowski MF, Lavin M, Sanderson MJ (2004) A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot 91:1846–1862
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinform 20:3252–3255
Zhang X, Wessler SR (2004) Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc Natl Acad Sci USA 101:5589–5594
Acknowledgments
This work was supported by Grant DEB–0120709 from the National Science Foundation to R.K.J. and J.L.B. Part of this work was performed under the auspices of the U.S. Department of Energy, Office of Biological and Environmental Research, by the University of California, Lawrence Berkeley National Laboratory, under contract DE-AC02-05CH11231. We thank Jeff Palmer for providing the purified plastid DNA used in this study and Stephen Downie and two anonymous reviewers for valuable comments on an early draft of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cai, Z., Guisinger, M., Kim, HG. et al. Extensive Reorganization of the Plastid Genome of Trifolium subterraneum (Fabaceae) Is Associated with Numerous Repeated Sequences and Novel DNA Insertions. J Mol Evol 67, 696–704 (2008). https://doi.org/10.1007/s00239-008-9180-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9180-7