Abstract
Differential display was used to isolate cDNA clones showing differential expression in response to ABA, drought and cold in barley seedling shoots. One drought-regulated cDNA clone (DD12) was further analyzed and found to encode a branched-chain amino acid aminotransferase (HvBCAT-1). A genomic clone was isolated by probing the Morex BAC library with the cDNA clone DD12 and the structure of Hvbcat-1 was elucidated. The coding region is interrupted by six introns and contains a predicted mitochondrial transit peptide. Hvbcat1 was mapped to chromosome 4H. A comparison was made to rice and Arabidopsis genes to identify conserved structural patterns. Complementation of a yeast (Saccharomyces cerevisiae) double knockout strain revealed that HvBCAT-1 can function as the mitochondrial (catabolic) BCATs in vivo. Transcript levels of Hvbcat-1, increased in response to drought stress. As the first enzyme in the branched-chain amino acid (BCAA) catabolic pathway, HvBCAT-1 might have a role in the degradation of BCAA. Degradation of BCAA could serve as a detoxification mechanism that maintains the pool of free branched-chain amino acids at low and non toxic levels, under drought stress conditions.
Similar content being viewed by others
Introduction
Plants have evolved adaptive strategies to cope with environmental stresses. Among the abiotic stresses, water availability is the most common and important in determining plant survival. Plants react to drought stress in a complex manner. The “adaptation syndrome” begins with stress perception, which initiates signal transduction pathways that cause changes at the cellular, physiological and developmental levels, which involve hormone balance, modification of metabolites and repression or induction of many regulated genes (Blum 1996).
Amino acid pools and concentration are influenced by abiotic stress: free amino acid levels are affected by increased protein degradation (Di Martino et al. 2003; Kaplan et al. 2004) and some amino acids act as compatible solutes (i.e. proline) whose synthesis is strongly induced by drought. Amino acids can act in defence mechanisms, for example the branched chain amino acids (BCAA) leucine, isoleucine, and valine serve as precursors of secondary metabolites involved in pathogen response, and their level is increased in response to drought and heat stress (Rizhsky et al. 2004).
Pathways for the biosynthesis and degradation of BCAA are well known in plants and many genes have been characterised (Singh 1999) with the exception of the final biosynthetic step, which is catalysed by a branched-chain amino acid transaminase (BCAT; EC 2.6.1.42). BCATs catalyse the final transamination step in the pathway that produces the essential amino acids leucine, isoleucine, and valine; the enzymes are also involved in the degradation of the three amino acids. BCAT activity has been detected in a wide range of eukaryotic organisms, including yeast, maize, spinach, pea, barley, tomato, soybean, rat and human (Singh 1999). Biosynthesis of BCAA in plants is localised in plastids, while there is evidence for a mitochondrial catabolic pathway (Diebold et al. 2002; Fujiki et al. 2000). After a transamination step, the α-keto-acids are esterified by the branched-chain α-keto-acid dehydrogenase complex (BCKDC) whose activity has been detected in Arabidopsis mitochondria. Several enzymes in the BCAA degradative pathway have also been localised in mitochondria by LC–MS/MS analysis of mitochondrial trypsinated proteins (Taylor et al. 2004).
Plants contain a small family of bcat genes. So far the identification and cloning of bcat genes in potato have demonstrated that the two genes isolated are differentially expressed (Campbell et al. 2001), whereas in Arabidopsis a family of six transcribed members with different sub-cellular localization has been described (Diebold et al. 2002). The isoenzyme AtBCAT-1 is imported into mitochondria while the other isoenzymes are either sorted into plastids or remain in the cytosol. It has been demonstrated that AtBCAT-1 can initiate the degradation of all branched-chain amino acids (Schuster and Binder 2005), thus confirming that plant mitochondria are the reaction compartment for the degradation of all BCAA.
Here we report the barley gene Hvbcat-1, which belongs to the BCAT gene family, regulated by drought stress. The coding region is interrupted by six introns and contains a predicted mitochondrial transit peptide. The gene was mapped to chromosome 4H. We propose that an increased degradation of BCAA under stress conditions could provide an alternative carbon source or detoxification mechanism by maintaining the pool of free branched-chain amino acids at levels compatible with cellular homeostasis.
Materials and methods
Plant material
Spring barley seeds (Hordeum vulgare L. cv. Georgie) were obtained from the Experimental Institute for Cereal Research, Fiorenzuola d’Arda (Italy). Seeds were surface-sterilized with 2% sodium hypochlorite for 10 min, rinsed with water, then germinated on moist filter paper and grown at room temperature (RT) in the dark for 3–4 days. Seedlings with approximately 3 cm coleoptiles were treated or stressed with ABA, NaCl, drought, cold and heat stress as described by Malatrasi et al. (2002). For microarray expression analysis plants were treated as described by Rodriguez et al. (2005).
The Oregon Wolfe Barley mapping population that contains 94 F1-derived doubled-haploid lines (http://www.barleyworld.org/oregonwolfebarleys/concept.php; Costa et al. 2001) was obtained from Dr. Patrick Hayes (Oregon State University) and propagated at the University of California, Riverside. Leaves and shoots were cut off, frozen in liquid nitrogen, and stored at −80°C until use.
Differential display reverse transcriptase and cloning
The differential display method (Liang and Pardee 1992) was performed with RNAmap kit (GenHunter, Nashville, TN) as directed by the manufacturer with the T12MN anchored primer and an arbitrary 10-mer primer (5′-GACCGCTTGT-3′). After re-amplification, the resulting fragments were separated on a 1.5% agarose gel and purified. These fragments were cloned in pGEM-T vector (Promega, Madison, WI) and sequenced.
Progressive drought stress, RWC and OP evaluation
Barley plants cv. Georgie were grown in a greenhouse at 23°C with 16 h of light (PPFD 80 μmol m−1 s−1) for 7 days with continuous watering, then irrigation was suspended for 2 weeks. Two pots with plants regularly watered for the entire period were used as control. After this period all pots were watered again and plants were sampled 2 h later. Leaf relative water content (RWC) was measured, according to Barr and Weatherley (1962), taking the first leaf of five plants after 7, 10, 11 and 14 days of water deprivation. Cellular osmotic potential (ψs) was measured on the first leaf of three plants after 7, 10, 11 and 14 days of water deprivation: leaves were sealed into a syringe and frozen immediately, the samples were then allowed to thaw at room temperature and the cell sap was collected in vials by pressing the tissues. The osmotic potential of the cell sap was measured using a vapour pressure osmometer (model 5500 Wescor, Logan, UT). Sub-samples were collected at the same time points for RNA purification.
Expression analysis by northern blot and real-time RT-PCR
Total RNA was extracted from 500 mg of seedlings (cv. Georgie) using a procedure adapted from Verwoerd et al. (1989). For northern blot analysis, 30 μg of RNA were separated on a denaturing formaldehyde 1.5% agarose gel and blotted onto Hybond N+ nylon membrane (Amersham Biosciences, Uppsala, Sweden) and hybridised according to Sambrook et al. (1989). The hybridisation signal was quantified by electronic autoradiography with the instrument Instant Imager (Canberra Packard, Meriden, CT). An equal quantity of RNA in each lane was verified by ethidium bromide staining using as a reference the intensity of rRNA bands.
For reverse transcriptase-PCR (RT-PCR) total RNA was treated with DNaseI (Promega) according to the manufacturer then 200 ng were retro-transcribed with 3′ gene specific primers using M-MLV point mutant-RNaseH minus (Promega). An aliquot of these reactions was used as template for PCR amplifications: reaction conditions were 95°C for 2 min followed by 40 amplification cycles with 30 s at 95°C for denaturation step, 30 s at 59°C for annealing and 15 s at 72°C for extension.
For real-time RT-PCR analysis, 500 ng of total RNA were retro-transcribed with random hexamers, then 20 ng used for PCR amplifications with DD12 specific primers (RT-DD12Forw 5′-GCGCCAGAAGCATCCAGTATT-3′; RT-DD12Rev 5′-CGATTTCTTTCCGTGCCCA-3′); 18S rRNA was also amplified as endogenous control (RT-18S Forw 5′-TGCCCTATCAACTTTCGATGG-3′; RT-18S Rev 5′- TGGATGTGGTAGCCGTTTCTC-3′). The reactions were performed in triplicate using Platinum SYBR Green qPCR Supermix UDG (Invitrogen, Carlsbad, CA). For both genes a calibration curve was calculated and the expression of DD12 normalized on the 18S rRNA.
Expression analysis by microarray
Expression data from triplicated experiments were collected using the Affymetrix Barley1 GeneChip (Affymetrix, Santa Clara, CA) and viewed using the software GeneSpring (Silicon Genetics, Redwood City, CA). Expression estimates were calculated using gcRMA implemented in GeneSpring 7.1 (Silicon Genetics) and normalization was done to the median of each probe set. Hvbcat-1 expression levels were studied using the probe set contig8247_at.
Genomic DNA isolation and genotyping
Genomic DNA of parental and doubled haploid genotypes was prepared from leaves using Plant DNAzol (Invitrogen) according to the instructions from the manufacturer. Gene-specific oligonucleotides (forward 5′-CTCCTCCTGCAACCTCTTCG-3′ and reverse 5′-CCGTGCAGAACACTTCGTCT-3′) were designed in order to amplify a genomic region containing a single nucleotide polymorphism between OWB-D and OWB-R forming in the latter a ClaI restriction site. PCR amplifications were performed with a GeneAmp PCR system 9600 (Perkin Elmer, Foster City, CA, USA) in a 40 μl reaction containing Taq DNA polymerase (Qiagen, Hilden, Germany), PCR buffer (10 mM Tris–HCl pH 8.3, 50 mM KCl2, 1.5 mM MgCl2), Q-solution, 200 μM of each dNTP, 0.3 μM primer, and 60–100 ng genomic DNA. PCR reactions were initiated at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 59°C for 30 s, 72°C for 45 s, and terminated at 72°C for 10 min. Amplified DNAs were digested with ClaI restriction enzyme and electrophoresed in a 2% MetaPhor® agarose gel (Cambrex, Baltimore, MD).
Linkage analysis
The genotype of each individual was recorded and used in Map Manager QTX software (Manly et al. 2001) and gene recombination frequencies were used to map Hvbcat-1. The Kosambi (1944) mapping function was used. The reference data set was available at http://www.barleyworld.org/oregonwolfebarleys/maps.php.
Isolation and characterization of the BAC clone
A 6.3× Morex barley BAC library 17-filter set (Yu et al. 2000) was obtained from Clemson University Genomics Institute. Pre-hybridization and hybridization conditions were described in Malatrasi et al. (2002). The DD12 cDNA fragment of Georgie barley was labelled with 32P-dCTP. DNA from a positive BAC clone (461O20) was isolated by an alkaline-lysis procedure (Sambrook et al. 1989) from a 20 ml overnight culture containing 12.5 μg/ml of chloramphenicol. BamHI and HindIII digestion fragments were separated in a 0.7% agarose gel, then blotted onto a nylon membrane and probed with [32P]-labeled DD12 cDNA following standard procedures (Sambrook et al. 1989). A 4 kb HindIII–EcoRI fragment of the BAC clone was subcloned in pUC19. Sequencing of the region containing Hvbcat-1 was performed throughout both strands using CEQ 2000 Dye Terminator Cycle Sequencing Quick Start Kit (Beckman Coulter, Fullerton, CA). A set of DD12 gene-specific primers was designed and verified with the program NetPrimer. Oligonucleotides were synthesized by MWG Biotech (Ebersberg, Germany).
The nucleotide and deduced amino acid sequences were analyzed and compared with sequences in databases using BLAST (Altschul et al. 1997) and ClustalW (Higgins et al. 1994) programs. Prediction of subcellular targeting was done using MitoProt II (http://ihg.gsf.de/ihg/mitoprot.html), ChloroP 1.1 (http://www.cbs.dtu.dk/services/ChloroP), iPSORT (http://hc.ims.u-tokyo.ac.jp/iPSORT/), TargetP 1.0 (http://www.cbs.dtu.dk/services/TargetP), SubLoc1.0 (http://www.bioinfo.tsinghua.edu.cn/SubLoc/) and Predotar 1.03 (http://genoplante-info.infobiogen.fr/predotar). Analysis of the promoter sequence was performed on 1,000 bp upstream of the translation start codon, using the software Signal Scan and a plant cis-acting regulatory DNA elements database (Higo et al. 1999). The putative transcription initiation site was recognized using the program at http://www.fruitfly.org/seq_tools/promoter.html.
Yeast (Saccharomyces cerevisiae) complementation analysis
For the complementation tests, Hvbcat-1 full length cDNA was amplified with primer pairs BglII–BcatF (5′-AGATCTATGGCTGTGCTGTCTTCTGCG-3′) and EcoRI–BcatR (5′-GAATTCTCAATCAATCGCTACCGTCC-3′) containing the BglII–EcoRI restriction sites at the 5′ ends to allow directional cloning into the yeast expression vector pYeDP10 under the control of phosphoglycerate kinase (PGK) promoter. To test the influence of sub-cellular targeting of HvBCAT-1 on functional complementation, primer pairs BglII–Bat1F (5′-AGATCTATGTTGCAGAGACATTCCTTG-3′) and BglII–Bat1 TP (5′-GGATCCTGCATCTAATGGGGCACCAG-3′) were used to amplify a fragment containing the targeting signal of S. cerevisiae BAT1 (aminoacid 1–25), which was then cloned upstream of the Hvbcat-1 full length cDNA (aminoacid 1–397) and upstream of the coding region from amino acids 42 trough 397 of HvBCAT-1 amplified using primer pairs BglII–41BcatF (5′-AGATCTATGAAGTCGTCGCTGCCGCAG-3′) and EcoRI–BcatR. The S. cerevisiae BAT1 gene was amplified using primer pairs BglII–Bat1F and EcoRI–BcatR (5′-GAATTCTCAATCAATCGCTACCGTCC-3′) and cloned in pYeDP10 and used as a positive control; the same vector without insert was used as a negative control. Escherichia coli DH5α was employed for plasmid DNA propagation. About 1 μg of each complementation clone and vector without insert were transformed into yeast strain Δbat2/gal–bat1. Yeast strain Δbat2/gal-bat1 (W303-A, bat2::LEU2; pBAT1::pGal1-10-HIS3) (Kispal et al. 1996) is a regulatory mutant that carries BAT1 under the control of an inducible Gal1-10 promoter and a deleted BAT2 gene. Deletions were carried out by a PCR-based method employing the LEU2-selective marker. Exchange of the endogenous promoter of the BAT1 gene for a galactose-inducible promoter in Δbat2 cells was performed as described (Lafontaine and Tollervey 1996). The Δbat2/gal-bat1 strain is not able to grow in absence of BCAA on minimal medium with glucose, while with galactose, the bat1 gene is expressed and the strain phenotype is essentially wild-type. The pYeDP10 vector, which carries the URA3 marker, rescues the auxotrophy for uracil. Cells were routinely grown in YPD medium (1% yeast extract, 2% Bacto peptone and 2% glucose). Yeast cells were transformed by the lithium chloride procedure (Ito et al. 1983), and transformants were selected and maintained in minimal medium containing 0.67% yeast nitrogen base without amino acids (Difco, Franklin Lakes, NJ) containing 2% (w/v) glucose and supplemented with adenine, lysine (20 mg/l each) and leucine (30 mg/l). Solid media were obtained with addition of 20 g/l agar; growth of yeast on solid media was observable after incubation at 28°C for 4 days.
Results
Isolation and characterisation of Hvbcat-1 in barley
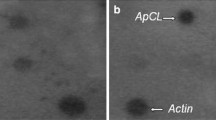
Differential display reverse transcription-PCR (DDRT-PCR) (Liang and Pardee 1992) was used for the amplification and isolation of partial cDNA fragments corresponding to stress-responsive genes as described in Malatrasi et al. (2002). In this work we focus our attention on one partial cDNA, DD12 (AJ493167), which is a 264 bp fragment. Expression of DD12 was determined in 3 days old seedlings by northern blot (Fig. 1) and real-time RT-PCR (data not shown). The transcript homologous to DD12 with a size of 1.6 kb was present in control conditions and did not significantly increase after treatment with ABA, NaCl, and in response to cold or heat stress. Under water deficit conditions a threefold increase was found and confirmed by real-time RT-PCR (data not shown).
Expression analysis of Hvbcat−1 in barley coleoptiles upon abiotic stresses and ABA treatment by northern blot. (C) Control; (ABA) incubation in abscisic acid; (D) drought stressed; (HS) heat stressed; (CS) cold treatment; (NaCl) incubation in NaCl solution. Total RNAs were isolated from coleoptiles of 3-day-old seedlings. Total RNA (30 μg) were separated by agarose gel electrophoresis, blotted and hybridised with DD12 cDNA fragment. Ethidium bromide-stained rRNA bands are shown on the bottom
The DD12 nucleotide sequence, as well as the translated sequence, were compared with nucleotide and protein sequences of the GenBank non-redundant (GenBank, Rockville Pike, Bethesda, MD) and expressed sequence tag (EST) databases by using BLAST (Altschul et al. 1997). A nucleotide–nucleotide BLASTN search revealed 10 ESTs from leaf cDNA library of barley almost identical to DD12 (95–100% of homology), while translation of the first 60 bp of DD12 5′ end showed significant homology with a putative branched-chain amino acid aminotransferase from Oryza sativa (AC135157-6) (data not shown). In order to obtain more sequence information to better characterize this gene (designated throughout this paper Hvbcat-1) the software HarvEST:Barley (http://www.harvest.ucr.edu/) was utilized. The consensus sequence of contig #8247 (assembly #21 unigene 21_8247) is 1,529 bp; this unigene corresponds to a transcript for a putative branched-chain amino acid aminotransferase of barley. The cDNAs comprising this contig derive mainly from drought stressed 3-week-old and normal 2-week-old seedling libraries. It was also possible to identify two other putative Hvbcat unigenes, 21_8040 and 21_20424, named throughout this paper Hvbcat-2 and Hvbcat-3, respectively. Hvbcat-2 is 1,690 bp and contains a complete open reading frame (ORF), Hvbcat-3 is 1,108 bp and lacks the 5′ region of the transcript.
The DD12 clone was used to probe a subset of the Morex BAC library filter set (equal to 1.1× coverage), resulting in the isolation of one BAC clone (461O20) containing the corresponding gene (Hvbcat-1). Comparison of the genomic DNA sequence of Hvbcat-1 with unigene 21_8247 showed the presence of six introns. The Hvbcat-1 gene has an ORF of 1,191 bp corresponding to a protein of 397 amino acids, with a deduced molecular mass of 42.7 kDa. HvBCAT-1 had highest homology with a rice putative BCAT (AAO06962, 76% of identity, 82% of similarity) and Arabidopsis AtBCAT-3 (CAB93131, 61% identity, 76% similarity). The N-termini of these proteins, as shown in Fig. 2a, are characterized by high sequence variability, probably due to different signal peptides for sub cellular sorting. HvBCAT-1 was predicted to have a mitochondrial signal peptide.
a Alignment of the amino acid sequences of branched-chain amino acid aminotransferases from barley (CAE00460), rice (AAO06962) and Arabidopsis (CAB93131). The alignment was made using the ClustalW program. Gaps in the sequences are indicated by dashes. “*” means that the residues in that column are identical in all sequences in the alignment. “:” means that conserved substitutions are observed. “.” means that semi-conserved substitutions are observed. Vertical bars indicate intron position. b Comparison of barley Hvbcat-1 (AJ574850), rice OsBCAT (AC105927) and Arabidopsis AtBCAT-3 (AJ276124) genomic clones. Exons positions are shown by thick rectangles and introns by thin rectangles. Four exons a, b, f, g are conserved in all sequences and e is conserved between barley and rice
Comparing Hvbcat-1 with the putative orthologs of rice and Arabidopsis (Fig. 2b) it is evident that these genes are formed by nine or ten exons, respectively. The position of introns is in most cases conserved, while their length is variable. Barley and rice have five exons conserved (a, b, e, f, g in Fig. 2b); barley and Arabidopsis have four (a, b, f, g) and the other exons have been subjected to rearrangements.
Analysis of the promoter region (1,000 bp) of Hvbcat-1 revealed a putative transcription initiation site at −138 bp and typical stress-related cis-acting elements including ABRE, DRE, MYC, MYB, AMYBOX and POLLEN1LELAT52 putative recognition sites (Table 1). The presence of these cis elements is consistent with the expression data.
Hvbcat-1 gene mapping in barley
Hvbcat-1 was mapped using a doubled haploid mapping population (Oregon Wolfe Barley; Costa et al. 2001) to barley chromosome 4H close to the centromere, it falls 2.2 cM from ×4.2 and 4.3 cM from Bmag0353 (Fig. 3). Chromosome 4H is known to contain some QTLs related to drought stress response (Teulat et al. 2001, 2003) including osmotic adjustment, RWC and water soluble carbohydrate, but the position of these QTL did not overlap with the map position of Hvbcat-1.
Genetic mapping of Hvbcat-1. Linkage map of chromosome 4H with the short arm at the top from the Oregon Wolfe Barley mapping population, based on Costa et al. (2001). Map location of Hvbcat-1 is underlined. Numbers on the left indicate the distance in centiMorgans; symbols on the right represent genetic markers
Hvbcat-1 expression is subjected to environmental control
Hvbcat-1 transcript levels were analysed in barley coleoptiles by real-time RT-PCR. To characterize the regulation of Hvbcat-1 under drought stress condition, a progressive dehydration was applied as described in Materials and methods. Leaf RWC and cellular sap osmotic potential were measured: RWC decreased from 96% in control plants to 80% in wilted plants after 14 days of water deprivation, whereas osmoticum concentrations increased from 443 to 788 mmol/kg; 2 h after re-watering RWC returned to levels of the controls whereas the osmotic potential remained significantly lower (Fig. 4a). Hvbcat-1 expression was evaluated in the same conditions by real-time RT-PCR (Fig. 4b): Hvbcat-1 was expressed at low levels in control conditions and expression increased sevenfold after 14 days of water deprivation. After 2 h re-watering the expression was fivefold over control samples.
Physiological and molecular analysis of progressive drought stress effects. (a) Time course of relative water content (RWC) and osmotic potential of barley plants under drought stress. Barley seedlings (after 7 days of growth) were dehydrated for 14 days, followed by re-hydration for 2 h; RWC and cellular osmotic potential were measured on the first leaf of each plant. Data are shown as the means ± SE (n = 5 for RWC; n = 3 for Os.Pot.). (b) Time course of Hvbcat-1 transcription under progressive drought stress evaluated by real-time RT-PCR. Total RNAs (500 ng) were retro-transcribed with random hexamers then 20 ng of cDNA were amplified with DD12 (AJ493167) specific primers and 18S rRNA specific primers as endogenous control. Quantification is based on C t values that were normalised using the C t value corresponding to a barley housekeeping gene (18S rRNA). Samples are a pool of four plants that were examined in triplicate. Each value is the mean ± SD
Expression analysis by microarray
To further analyze the expression of Hvbcat-1 and the two other putative members of the barley bcat family (Hvbcat-2 and Hvbcat-3) we used data gathered using the Affymetrix Barley1 GeneChip (Close et al. 2004) from various stress experiments (E. Rodríguez and J. T. Svensson, unpublished) and from another study including 15 different tissues/stages of unstressed plants (A. Druka, unpublished). The probe set named contig8247_at represents the Hvbcat-1 gene, while probe sets contig8040_at and contig20424_at correspond to Hvbcat-2 and Hvbcat-3 (http://harvest.ucr.edu). Data analysis produced a present call for contig8247_at in all samples, consistent with Hvbcat-1 constitutive expression found by real-time RT-PCR. Microarray data showed a strong transcript increase after drought-stress, a progressive transcript increase over control samples was recorded with a maximum of 34-fold at SWC7. Comparison among tissues showed the highest differences; for example, Hvbcat-1 transcripts were 6 times higher in anthers and 12 times higher in leaf than in reference sample (Fig. 5). The Hvbcat-2 transcript was revealed in all conditions analysed, with no apparent regulation either by environmental stresses or by developmental conditions except in anthers were a reduction of threefold over control samples was observed. Hvbcat-3 transcript, corresponding to contig20424_at, was not up- or down-regulated by the environmental stresses considered. Analysis of the expression level among tissues of this contig evidenced an increase of 37-fold in immature inflorescence and 31-fold in anthers, of 91-fold in seminal roots and of 97-fold in roots.
Hvbcat-1 transcript detection using microarrays, and comparison with two putative BCAT of barley. Microarray data from a stress experiment and b different tissues and development stages (A. Druka unpublished; E. Rodriguez, and J. T. Svensson, unpublished). Data were analyzed using GeneSpring, expression values were normalized to the median of each probe set. Signal intensity is the normalized probe-set fluorescence in arbitrary units. Transcript levels of Hvbcat-1 (contig 8247_at) are compared with transcript levels of contig 8040_at (Hvbcat-2) and contig 20424_at (Hvbcat-3) of barley
HvBCAT-1 function as BCATs in yeast
To determine the function of HvBCAT-1 an in vivo complementation test was performed using a Δbat2/gal-bat1 yeast strain (derivative of Kispal et al. 1996). This strain is unable to grow on minimal medium with glucose in the absence of a single, any combination of two, or all three branched-chain amino acids. Several transformation cassettes were prepared in the pYeDP10 vector, containing Scbat1 and Hvbcat-1 ORF downstream a PGK promoter, and used to transform Δbat2/gal-bat1 yeast strain (Fig. 6a). No complementation was achieved with the Hvbcat-1 full length cDNA (construct 1 Fig. 6a) but growth restoration on minimal medium lacking Ile and Val was obtained for the positive control (Fig. 6b-2). This led to the interpretation that either the gene Hvbcat-1 does not encode a BCAT or that the protein, predicted as mitochondrial, remained in the cytosol where it is not able to function. To check whether an improper sub-cellular targeting might be responsible for the lack of complementation, the predicted HvBCAT-1 mitochondrial transit peptide (amino acids 1–41) was removed (construct 3 Fig. 6a) and the sequence without the portion coding for the signal peptide or the full-length cDNA were fused to a predicted ScBAT1 mitochondrial targeting sequence (amino acids 1–25, constructs 4 and 5; Fig. 6a). Transformation of the strain Δbat2/gal-bat1 with these new constructs resulted in positive complementation for both constructs (4 and 5), demonstrating that the gene Hvbcat-1 from barley rescued the auxotrophy and therefore encodes a branched-chain amino acid aminotransferase (Fig. 6b).
Complementation analysis of HvBCAT-1 in the yeast double knockout mutant Δbat1/galbat2. To examine the suggested BCAT function in vivo, Hvbcat-1 cDNA was cloned into pYeDP10 vector and transformed into Δbat1/galbat2. The constructs prepared are shown in panel (a) 1- the complete ORF of Hvbcat-1 is cloned downstream the PGK promoter; 2-yeast BAT1 is used as positive control; 3-Hvbcat-1 ORF is deleted in 5′ of 123 bp coding for the transit peptide (41 aa); 4-BAT1 transit peptide is cloned in 5′ in frame with Hvbcat-1 deleted of 123 bp; 5-BAT1 transit peptide is cloned in 5′ in frame with complete Hvbcat-1 ORF; 6-pYeDP10 without insert is used as negative control. (b) Restored growth on medium lacking Val, Leu and Ile due to construct 4 and 5 indicates that the competent BCAT activity of HvBCAT-1 in yeast depends on the presence a mitochondrial transit peptide recognized by yeast. Complementation is also observed with ScBAT-1 as positive control (2). No growth is detected with the empty transformation vector (6) and with HvBCAT-1 with its own transit peptide (1) or without any transit peptide (3)
Discussion
In this work we describe the isolation, sequence, expression patterns, genetic map location and in vivo complementation test of a gene named Hvbcat-1 encoding a branched-chain amino acid aminotransferase regulated during some environmental conditions.
The Hvbcat-1 gene is composed of seven exons and encodes a 397 amino acid BCAT with a predicted mitochondrial localisation. BCATs catalyse the final transamination step in the pathway that produces leucine, isoleucine, and valine; these enzymes are also involved in the degradation of these three amino acids. The biosynthetic and the catabolic pathways are localized in different sub-cellular compartments with the former in plastids and the latter probably in mitochondria. To predict protein sub-cellular sorting of HvBCAT-1 we used different programs (iPSORT, TargetP, Predotar, MitoProt, SubLoc) and a mitochondrial localisation of HvBCAT-1 was suggested, indicating an involvement of this enzyme in BCAA degradation. The compartments where branched-chain amino acids are degraded in plants have been a matter of debate for many years. There is evidence that it occurs in peroxisomes (Gerbling and Gerhardt 1989), but other evidence assigned the β-oxidation of branched-chain α-keto acids to mitochondria, supported by the isolation and identification in Arabidopsis of mitochondrial proteins corresponding to several enzymes involved in this pathway, including BCAT (Fujiki et al. 2000; Diebold et al. 2002; Taylor et al. 2004). In particular it has been shown that the mitochondrial AtBCAT-1 can initiate the degradation of all BCAA in almost all tissues (Schuster and Binder 2005). The Hvbcat-1 gene encodes for a functional BCAT enzyme as shown by yeast complementation analysis. The yeast complementation experiments are consistent with mitochondrial targeting of HvBCAT-1 therefore, we consider it functionally homologous to the Arabidopsis enzyme, although the highest sequence homology was observed with AtBCAT-3 (Fig. 2a).
The analysis of Hvbcat-1 expression was performed in barley in several environmental and developmental conditions with northern analysis and quantitative real-time PCR (cv. Georgie) and with microarrays (cv. Morex). Northern analysis demonstrated that Hvbcat-1 transcript increased only in response to drought (Fig. 1) as confirmed also by real time RT-PCR (data not shown), therefore a more detailed analysis was performed. The experiment of progressive dehydration demonstrated that Hvbcat-1 transcription was increased in conditions of low RWC and low osmotic potential (Fig. 4). Similar expression patterns were confirmed with microarray data in cv. Morex in which there was a peak of expression at the lower SWC (soil water content) point (Fig. 5a). Furthermore, it was shown that this gene is actively transcribed in leaves, seedlings and anthers. The expression of Hvbcat-1 correlates with the localisation of Atbcat-1 by histo-chemical staining (Schuster and Binder 2005). The level of transcripts in cv Georgie was higher in coleoptiles as compared to leaves (data not shown), while in cv. Morex higher levels of Hvbcat-1 transcripts were evidenced in leaves. This discrepancy can be explained by different behaviour of the two cultivars or by the different growth stage of the coleoptiles analysed; the samples analysed by real-time RT-PCR were obtained from 3 cm long coleoptiles while in the array experiment the sampled coleoptiles were 2 mm long.
By a TBLASTN search of the EST database with HvBCAT-1, the presence of at least three genes seems possible in barley, as shown also using the HarvEST database. As is typical of multigene families, transcriptional analysis of three paralogous bcat genes by microarray shows that the three genes are not co-ordinately regulated. Similarly in Arabidopsis distinct expression patterns for the six transcribed members of the Atbcat gene family were observed (Schuster and Binder 2005), thus suggesting that each protein performs a specific function in different organs and tissues.
In the catabolic pathway of branched-chain amino acids, BCAT represents the first enzyme, followed by the branched-chain α-keto-acid dehydrogenase complex (BCKDC); in Arabidopsis it has been shown that genes encoding for BCKDC complex are induced by darkness, mannitol, and sugar starvation (Fujiki et al. 2002). Taylor and co-authors (2004) suggest that branched-chain amino acids promote their own catabolism but only when plant cells are sugar starved, and they may constitute an alternative source of respiratory substrates for the TCA cycle during severe plant stress. The results shown in Fig. 4 are consistent with this hypothesis, in fact it is well known that as RWC decreases a decline in photosynthetic rate is observed and an increased degradation of BCAA can serve as an alternative energy source. Hvbcat-1 is up-regulated also in etiolated coleoptile; this could be related to the progressive degradation of starch reserves of the endosperm and is consistent with the presence of specific cis-elements in its promoter, required for gene expression during sugar starvation.
BCAA and α-keto-acids are also considered to be cytotoxic, and can induce apoptosis in mammals (Eden and Benvenisty 1996). Thus, removal of BCAA via respiratory oxidation could also serve as a cell detoxification mechanism, maintaining a pool of branched chain amino acids for protein synthesis and at the same time preventing toxic levels. In stress conditions, such as those imposed to barley plants in this work, rapid protein degradation and accumulation of certain amino acids (e.g. proline) was observed (Reddy et al. 2004; Lawlor and Cornic 2002), BCAA level increased and therefore activation of their catabolism may play an important role as detoxification mechanism which may protect plant cells.
In Arabidopsis it has been observed that Atbcat-1 can use as a substrate also methionine and the corresponding α-keto-acids, suggesting an involvement of this enzyme in the methionine regeneration pathway (Berger et al. 2003). Methionine, besides being essential for protein synthesis, has an important role in stress response. In fact, it is mainly presents in cell as S-adenosine-methionine (SAM), which is utilised as a substrate in methylation reactions (e.g. choline) and as precursor in the biosynthesis of ethylene and polyamines. Therefore a further role for BCATs can be envisaged in relation to stress response.
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (HLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 2:3389–3402
Barr HD, Weatherley PE (1962) A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust J Biol Sci 1515:413–428
Bate N, Twell D (1998) Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol Biol 37:859–869
Berger BJ, English S, Chan G, Knodel MH (2003) Methionine regeneration and aminotransferase in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis. J Bacteriol 185:2418–2431
Blum A (1996) Crop responses to drought and the interpretation of adaptation. Plant Growth Regul 20:135–148
Busk PK, Jensen AB, Pages M (1997) Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. Plant J 11:1285–1295
Campbell MA, Patel JK, Meyers JL, Myrick LC, Gustin JL (2001) Genes encoding for branched-chain amino acid aminotransferase are differentially expressed in plants. Plant Physiol Biochem 39:855–860
Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise RP (2004) A new resource for cereal genomics: 22 K barley GeneChip comes of age. Plant Physiol 134:960–968
Costa JM, Corey A, Hayes PM, Jobet C, Kleinhofs A, Kopisch-Obusch A, Kramer SF, Kudrna D, Li M, Riera-Lizarazu O, Sato K, Szucs P, Toojinda T, Vales MI, Wolfe RI (2001) Molecular mapping of the Oregon Wolfe Barleys: a phenotypically polymorphic doubled-haploid population. Theor Appl Genet 103:415–424
Diebold R, Schuster J, Daschner K, Binder S (2002) The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol 129:540–550
Di Martino C, Delfine S, Pizzuto R, Loreto F, Fuggi A (2003) Free amino acids and glycine in leaf osmoregulation of spinach responding to increasing salt stress. New Phytol 158:455–463
Dunn MA, White AJ, Vural S, Hughes MA (1998) Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.). Plant Mol Biol 38:551–564
Eden A, Benvenisty N (1999) Involvement of branched-chain amino acid aminotransferase (Bcat1/Eca39) in apoptosis. FEBS Lett 457:255–261
Fujiki Y, Sato T, Ito M, Watanabe A (2000) Isolation and chracterization of cDNA clones for E1β and E2 subunits of the branched-chain α-ketoacid deydrogense complex in Arabidopsis. J Biol Chem 275:6007–6013
Fujiki Y, Ito M, Itoh T, Nishida I, Watanabe A (2002) Activation of the promoters of Arabidopsis genes for the branched-chain a-keto acid dehydrogenase complex in transgenic tobacco BY-2 cells under sugar starvation. Plant Cell Physiol 43:275–280
Gerbling H, Gerhardt B (1989) Peroxisomal degradation of brached-chain 2-oxo-acids. Plant Physiol 91:1387–1392
Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database:1999. Nucleic Acids Res 27:297–300
Huang N, Sutliff TD, Litts JC, Rodriguez RL (1990) Classification and characterization of the rice alpha-amylase multigene family. Plant Mol Biol 14:655–668
Hwang YS, Karrer EE, Thomas BR, Chen L, Rodriguez RL (1998) Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation. Plant Mol Biol 36:331–341
Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153:163–168
Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller C, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature–stress metabolome of Arabidopsis. Plant Physiol 136:4159–4168
Kispal G, Steiner H, Court DA, Rolinski B, Lill R (1996) Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of myc oncogene-regulated Eca39 protein. J Biol Chem 271:24458–24464
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lafontaine D, Tollervey D (1996) One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acid Res 24:3469–3472
Lawlor DW, Cornic G (2002) Photosynthetic carbon assimilation and associated metabolism in relation to water deficit in higher plants. Plant Cell Environ 25:275–294
Liang P, Pardee AB (1992) Differential display of eukaryotic messenger RNA by means of polymerase chain reaction. Science 257:967–971
Malatrasi M, Close TJ, Marmiroli N (2002) Identification and mapping of a putative stress response regulator gene in barley. Plant Mol Biol 50:141–150
Manly K, Cudmore J, Meer J (2001) Map manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932
Reddy AR, Chaitanya KV, Vivekanandan M (2004) Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J Plant Physiol 161:1189–1202
Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134:1683–1696
Rodriguez EM, Svensson JT, Malatrai M, Choi D-W, Close TJ (2005) Barley Dhn13 encodes a KS-type dehydrin with costitutive and stress responsive expression. Theor Appl Genet 110:852–858
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Plainview, NY
Schuster J, Binder S (2005) The mitochondrial branched-chain aminotransferase (AtBCAT-1) is capable to initiate degradation of leucine, isoleucine and valine in almost all tissue of Arabidopsis thaliana. Plant Mol Biol 57:241–254
Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J 33:259–270
Singh BK (1999) Biosynthesis of valine, leucine and isoleucine. In: Singh BK (ed) Plant amino acids: biochemistry and biotechnology. Marcel Dekker, New York, pp 227–247
Taylor NL, Heazlewood JL, Day DA, Millar AH (2004) Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134:838–848
Teulat B, Borries C, This D (2001) New QTLs identified for plant water status, water-soluble carbohydrate and osmotic adjustement in a barley population grown in a growth-chamber under two water regimes. Theor Appl Genet 103:161–170
Teulat B, Zoumarou-Wallis N, Rotter B, Ben Salem M, Bahri H, This D (2003) QTL for relative water content in field-grown barley and their stability across Mediterranean environments. Theor Appl Genet 108:181–188
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539
Verwoerd TC, Dekker BMM, Hoekema A (1989) A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17:2362
Yu Y, Tomkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, Brueggeman RS, Muehlbauer GJ, Wise RP, Wing RA (2000) A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes. Theor Appl Genet 101:1093–1099
Acknowledgments
We are grateful to prof. R. Lill and U. Muehlenhoff (University of Munchen, Germany) for the generous gift of the Δbat2/gal-bat1 yeast strain and to prof. Tiziana Lodi (University of Parma, Italy) for the gift of the plasmid pYeDP10 for yeast transformation. This work has been supported by NATO Grant (CLG 978261) to N. Marmiroli, by project “Biotecnologie Vegetali” (MIPA) to N. Marmiroli, by CNR-Agenzia 2000 project to M. Gulli and in part by NSF DBI-0321756, “Coupling Expressed Sequences and Bacterial Artificial Chromosome Resources to Access the Barley Genome” to T.J. Close.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Langridge
M. Malatrasi and M. Corradi equally contributed to this research paper.
Rights and permissions
About this article
Cite this article
Malatrasi, M., Corradi, M., Svensson, J.T. et al. A branched-chain amino acid aminotransferase gene isolated from Hordeum vulgare is differentially regulated by drought stress. Theor Appl Genet 113, 965–976 (2006). https://doi.org/10.1007/s00122-006-0339-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0339-6