Abstract.
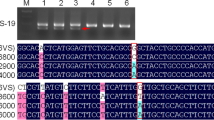
PCR with the DNA of translocation chromosomes and marker-specific primers has been used to merge genetically mapped microsatellite (MS) markers into the physically integrated restriction fragment length polymorphism (RFLP) map of barley chromosome 3H. It was shown that the pronounced clustering of MS markers around the centromeric region within the genetic map of this chromosome results from suppressed recombination. This yielded a refinement of the physically integrated RFLP map of chromosome 3H by subdivision of translocation breakpoints (TBs) that were previously not separated by markers. The physical distribution of MS markers within most of the subchromosomal regions corresponded well with that of the RFLP markers, indicating that both types of markers are similarly valuable for a wide range of applications in barley genetics.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Electronic Publication
Rights and permissions
About this article
Cite this article
Künzel, .G., Waugh, .R. Integration of microsatellite markers into the translocation-based physical RFLP map of barley chromosome 3H. Theor Appl Genet 105, 660–665 (2002). https://doi.org/10.1007/s00122-002-0913-5
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00122-002-0913-5