Abstract
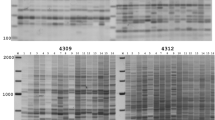
Rice is a model genome for cereal research, providing important information about genome structure and evolution. Retrotransposons are common components of grass genomes, showing activity at transcription, translation and integration levels. Their abundance and ability to transpose make them good potential markers. In this study, we used 2 multilocus PCR-based techniques that detect retrotransposon integration events in the genome: IRAP (inter-retrotransposon amplified polymorphism) and REMAP (retrotransposon-microsatellite amplified polymorphism). Markers derived fromTos17, a copia-like endogenous retrotransposon of rice, were used to identify genetic similarity among 51 rice cultivars (Oryza sativa L.). Genetic similarity analysis was performed by means of the Dice coefficient, and dendrograms were developed by using the average linkage distance method. A cophenetic correlation coefficient was also calculated. The clustering techniques revealed a good adjustment between matrices, with correlation coefficients of 0.74 and 0.80, or lower (0.21) but still significant, between IRAP and REMAP-based techniques. Consistent clusters were found for Japanese genotypes, while a subgroup clustered the irrigated Brazilian genotypes.
Similar content being viewed by others
References
Amar K, Hirochika H, 2001. Applications of retrotransposons as genetic tools in plant biology. Trends in Plant Sci 6: 127–133.
Briard M, Clera VLE, Gnzebelus D, Senalik D, Simon PW, 2000. Modified protocols for rapid carrot genomic DNA extraction and AFLP™ analysis using silver stain on radioisotopes. Plant Mol Biol Rep 18: 235–241.
Dice LR, 1945. Measures of the amount of ecological association between species. Ecology 26: 297–307.
Flavell RB, Rimpau J, Smith DB, 1977. Repeated sequence DNA relationships in four cereal genomes. Chromosoma 63: 205–222.
Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, et al. 2000.fw 2,2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88.
Garry AJ, Tai TH, Coburn J, Kresovich S, McCouch S, 2005. Genetic structure and diversity inOryza sativa L. Genetics 169: 1631–1638.
Hirochika H, 2001. Contribution of theTos17 retrotransposon to rice functional genomics. Curr Op Plant Biol 4: 118–122.
Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M, 1996. Retrotransposon of rice involved in mutation by culture. Proc Natl Acad Sci 93: 7783–7788.
[IRGSP] International Rice Genome Sequencing Project, 2005. The map-based sequence of the rice genome. Nature 436: 793–800.
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman I, 1999. IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98: 704–711.
Malone G, Zimmer PD, Mattos LAT, Kopp MM, Carvalho FIF, de Oliveira AC, 2006. Assessment of the genetic variability among rice cultivars revealed by AFLP. Revista Brasileira de Agrociencia 12: 21–25.
Mantel N, 1967. The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220.
McCouch SR, 2001. Genomics and synteny. Plant Physiol 125: 152–155.
Oliveira AC de, Richter T, Bennetzen, JL, 1996. Regional and racial specificities in sorghum germplasm assessed with DNA markers. Genome 39: 579–587.
Pandini F, Carvalho FIF, Barbosa Neto JF, 1997. Plant height reduction in populations of triticale (×Triticosecale Wittmack) by induced mutations and artificial crosses. Braz J Genet 20: 483–488.
Price Z, Schulman AH, Mayes S, 2003. Development of new marker methods — an example from oil palm. Plant Genetics Res 1: 103–113.
Rohlf FJ, 1972. An empirical comparison of three ordination techniques in numerical taxonomy. Systematic Zool 21: 271–280.
Rohlf FJ, 2000. NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.1. Exeter Software, New York.
SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, et al. 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768.
Sokal RR, Rohlf FJ, 1962. The comparison of dendrograms by objective methods. Taxon 11: 30–40.
Vitte C, Panaud O, 2005. LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenet Genome Res 110: 91–107.
Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BBT, Powell W, 1997. Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol Gen Genet 253: 687–694.
Yap IV, Nelson RJ, 1996. Winboot: a program for performing bootstrap analysis of binary data to determine the confidence limits of UPGMA-based dendrograms. Manila: IRRI.
Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Potrykus I, 2000. Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Branco, C.J.S., Vieira, E.A., Malone, G. et al. IRAP and REMAP assessments of genetic similarity in rice. J Appl Genet 48, 107–113 (2007). https://doi.org/10.1007/BF03194667
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03194667