Abstract
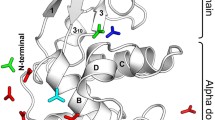
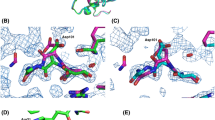
This paper summarizes our crystallographic studies of the interaction of denaturants with cross-linked triclinic lysozyme. Electron density maps of various bromoethanol-lysozyme complexes are analyzed and compared to those reported earlier for SDS-lysozyme complexes. Despite differences in the chemical nature and size of the two denaturants their mode of interaction with the protein is quite similar, suggesting the existence of a general mechanism for binding of hydrophobic-hydrophilic denaturants to proteins. Our results are consistent with the conclusion that lysozyme consists of two domains connected by a flexible segment and that this segment represents an internal degree of freedom of the protein.
Similar content being viewed by others
References
Tanford, C.: Protein denaturation. Adv. Protein Chem. 23, 121–282 (1968)
Tanford, C.: Protein denaturation. Adv. Protein Chem. 24, 1–95 (1970)
Yonath, A., Sielecki, A., Podjarny, A., Moult, J., Traub, W.: Studies of protein denaturation and renaturation. I. Effects of denaturation on lysozyme crystals. Biochemistry 16, 1413–1417 (1977)
Yonath, A., Podjarny, A., Honig, B., Sielecki, A., Traub, W.: Crystallographic studies of protein denaturation and renaturation. II. SDS induced structural changes in triclinic lysozyme. Biochemistry 16, 1418–1423 (1977)
North, A. C. T., Phillips, D. C., Mathews, F. S.: A semi-empirical method of absorption correction. Acta Cryst. A24, 351–359 (1968)
Wilson, A. J. C.: Determination of absolute from relative X-ray intensity data. Nature (Lond.) 150, 151–152 (1942)
Moult, J., Yonath, A., Traub, W., Smilansky, A., Podjarny, A., Saya, A., Rabinovich, D.: The structure of triclinic lysozyme at 2.5 å resolution. J. Mol. Biol. 100, 179–195 (1976)
Imoto, T., Johnson, L. N., North, A. C. T., Phillips, D. C., Rupley, J. A.: In: The enzymes, vol. VII, 3rd ed. (ed. P. D. Boyer), pp. 665–686. New York: Academic Press 1972
Rupley, J. A., Gates, V., Bilbrey, R.: Lysozyme catalysis, evidence for carbonium ion intermediate and participation of glutamic acid 35. J. Amer. Chem. Soc. 90, 5633 (1968)
Vincentelli, J. B., Looze, Y., Leonis, J.: Etude de l'effect denaturant d'alcools polyhydroxyliques sur le lysozyme à l'état dissous. Arch. Int. Physiol. Biochim. 79, 855 (1971)
Author information
Authors and Affiliations
Additional information
The work was carried out during the tenure of a fellowship from the European Molecular Biology Organization
We are grateful to Dr. Gerson Cohen for providing us with his data processing programs, to Drs. David Haas, Paul Sigler, Thomas Creighton and Micael James for helpful discussions, and to Mr. Samuel Getteno for his invaluable technical assistance.
Rights and permissions
About this article
Cite this article
Yonath, A., Podjarny, A., Honig, B. et al. Structural analysis of denaturant-protein interactions: Comparison between the effects of bromoethanol and SDS on denaturation and renaturation of triclinic lysozyme. Biophys. Struct. Mechanism 4, 27–36 (1978). https://doi.org/10.1007/BF00538838
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00538838