Summary
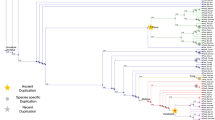
The Kunitz-type protease inhibitor is one of the serine protease inhibitors. It is found in blood, saliva, and all tissues in mammals. Recently, a Kunitz-type sequence was found in the protein sequence of the amyloid β precursor protein (βAPP). It is known that βAPP accumulates in the neuritic plaques and cerebrovascular deposits of patients with Alzheimer's disease. Collagen type VI in chicken also has an insertion of a Kunitz-type sequence. To elucidate the evolutionary origin of these insertion sequences, we constructed a phylogenetic tree by use of all the available sequences of Kunitz-type inhibitors. The tree shows that the ancestral gene of the Kunitz-type inhibitor appeared about 500 million years ago. Thereafter, this gene duplicated itself many times, and some of the duplicates were inserted into other protein-coding genes. During this process, the Kunitz-type sequence in the present βAPP gene diverged from its ancestral gene about 270 million years ago and was inserted into the gene soon after duplication. Although the function of the insertion sequences is unknown, our molecular evolutionary analysis shows that these insertion sequences in βAPP have an evolutionarily close relationship with the inter-α-trypsin inhibitor or trypstatin, which inhibits the activity of tryptase, a novel membrane-bound serine protease in human T4+ lymphocytes.
Similar content being viewed by others
References
Bonaldo P, Russo V, Bucciotti F, Doliana R, Colombatti A (1990) Structural and functional features of the alpha-3 chain indicate a bridging role for chicken collagen VI in connective tissues. Biochemistry 29:1245–1254
Cechova D, Jonakova V, Sorm F (1971) Protein Identification Resource (PIR). National Biomedical Research Foundation, Washington DC
Creighton TE, Charles IG (1987) Sequences of the genes and polypeptide precursors for two bovine protease inhibitors. J Mol Biol 194:11–22
Dufton MJ (1985) Proteinase inhibitors and dendrotoxins. Sequence classification, structural prediction and structure/activity. Eur J Biochem 153:647–654
Fioretti E, Binotti I, Barra D, Citro G, Ascoli F, Anonini E (1983) Heterogeneity of the basic pancreatic inhibitor (Kunitz) in various bovine organs. Eur J Biochem 130:13–18
Moretti E, Iacopino G, Angreletti M, Barra D, Bossa F, Ascoli F (1985) Primary structure and antiproteolytic activity of a Kunitz-type inhibitor from bovine spleen. J Biol Chem 260: 11451–11455
Fukuchi K, Martin GM, Deeb SS (1989) Sequence of the protease inhibitor domain of the A4 amyloid protein precursor of Mus domesticus. Nucleic Acids Res 17:5396
Gifard TJ, Wesselschmidt RL, Broze GJ Jr (1991) cDNA sequence of rabbit lipoprotein-associated coagulation inhibitor. Genetic Sequence Data Bank (Genbank), CA
Gojobori T, Ishii K, Nei M (1982) Estimation of average number of nucleotide substitutions when the rate of substitution varies with nucleotide. J Mol Evol 18:414–423
Graur D, Li WH (1988) Evolution of protein inhibitors of serine proteinases: positive Darwinian selection or compositional effects? J Mol Evol 28:131–135
Hawkins RL, Seeds NW (1986) Effect of protease and their inhibitors on neurite outgrowth from neonatal mouse sensory ganglia in culture. Brain Res 398:63–70
Hill RE, Hastie ND (1987) Accelerated evolution in the reactive center regions of serine protease inhibitors. Nature 326: 96–99
Hochstrasser K, Wachter E, Albrecht GJ, Reisinger P (1985) Protein Identification Resource (PIR). National Biomedical Research Foundation, Washington DC
Hokama Y, Iwanaga S, Tatsuki T, Suzuki T (1976) Protein Identification Resource (PIR). National Biomedical Research Foundation, Washington DC
Ikeo K, Takahashi K, Gojobori T (1991) Evolutionary origin of numerous kringles in human and simian apolipoprotein (a). FEBS Lett 287:146–148
Javaherian K, Langlois AJ, McDanal C, Ross KL, Eckler LI, Jellis CL, Profy AT, Rusche JR, Bolognesi DP, Putney SD, Matthews TJ (1989) Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc Natl Acad Sci USA 86:6768–6772
Joubert FJ, Strydom DJ (1978) Protein Identification Resource (PIR). National Biomedical Research Foundation, Washington DC
Kang J, Mueller-Hill B (1989) The sequence of the two extra exons in rat preA4. Nucleic Acids Res 17:2130
Kassell B, Laskowski M (1965) The basic inhibitor of bovine pancreas V. The disulfide linkages. Biochem Biophys Res Commun 20:463–468
Kato I, Tominaga N (1979) Protein Identification Resource (PIR). National Biomedical Research Foundation, Washington DC
Kaumeyer JF, Polazzi JO, Kotick MP (1986) The mRNA for a proteinase inhibitor related to the HI-30 domain of interalpha-trypsin inhibitor also encodes alpha-1-microglobulin (protein HC). Nucleic Acids Res 14:7839–7850
Kido H, Yokogoshi Y, Katunuma N (1988) Kunitz-type protease inhibitor found in rat mast cells. Purification, properties, and amino acid sequence. J Biol Chem 263:18104–18107
Kido H, Fukutomi A, Katsunuma N (1991) Tryptase TL2 in the membrane of human T4+ lymphocytes is a novel binding protein of the V3 domain of HIV-1 envelope glycoprotein gpl20. FEBS Lett 287:233–236
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge, UK
Kitaguchi N, Takahashi Y, Oishi K, Shiojiri S, Tokushima Y, Utsunomiya T, Ito H (1990) Enzyme specificity of proteinase inhibitor region in amyloid precursor protein of Alzheimer's disease: different properties compared with protease nexin I. Biochim Biophys Acta 1038:105–113
Kondo K, Toda H, Narita K, Lee CY (1982) Protein Identification Resource (PIR). National Biomedical Research Foundation, Washington DC
Koo EH, Sisodia SS, Price DL (1989) The sequence of the monkey amyloid precursor protein protease inhibitor domain. Genetic Sequence Data Bank (Genbank), CA
Laskowski M, Kato I (1980) Protein inhibitors of proteinases. Annu Rev Biochem 49:593–626
Laskowski M, Kato I, Lery TR, Schrode J, Sealock RW (1974) Evolution of specificity of protein proteinase inhibitors. In: Proteinase inhibitors. Bayer-Symposium V, pp 597–611
Muller-Hill B, Beyreuther K (1989) Molecular biology of Alzheimer's disease. Annu Rev Biochem 58:287–307
Nakamura T, Hirai T, Tokunaga F, Kawabata S, Iwanaga S (1987) Purification and amino acid sequence of Kunitz-type protease inhibitor found in the hemocytes of horseshoe crab (Tachypleus tridentatus). J Biochem 101:1297–1306
Nei M (1975) Molecular population genetics and evolution. North-Holland, Amsterdam
Ponte P, Gonzalez-DeWhitt P, Schilling J, Miller J, Hsu D, Greenberg B, Davis K, Wallace W, Lieberburg I, Fuller F, Cordell B (1988) A new A4 amyloid mRNA contains a domain homologous to serine inhibitors. Nature 333:525–527
Ramesh N, Sugumaran M, Mole JE (1988) Purification and characterization of two trypsin inhibitors from the hemolymph of Manduca sexta larvae. J Biol Chem 263:11523–11527
Ritonja A, Meloun B, Gubensek F (1983) Protein Identification Resource (PIR). National Biomedical Research Foundation, Washington DC
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salvessen G, Nagase H (1989) Inhibitor of proteolytic enzymes In: Beynon RJ, Bond JS (eds) Proteolytic enzymes. IRL Press, UK, pp 83–104
Sasaki T (1984) Amino acid sequence of a novel Kunitz-type chymotrypsin inhibitor from hemolymph of silkworm larvae, Bombyx mori. FEBS Lett 168:227–230
Sasaki T (1988) Amino-acid sequences of two basic chymotrypsin inhibitors from silkworm larval hemolymph. Biol Chem Hoppe-Seyler 369:1235–1241
Schmidt T, Stumm-Zollinger E, Chen PS, Bohlen P, Stone SR (1989) A male gland peptide with protease inhibitory activity in Drosophila funebris. J Biol Chem 264:9745–9749
Siekmann J, Wenzel HR, Schroeder W, Tschesche H (1988) Characterization and sequence determination of six aprotinin homologues from bovine lungs. Biol Chem Hoppe-Seyler 369: 157–163
Strydom DJ (1973) Protease inhibitors as snake venom toxins. Nature New Biol 243:88–89
Strydom DJ, Joubert FJ (1981) Protein Identification Resource (PIR). National Biomedical Research Foundation, Washington DC
Takahashi H, Iwanaga S, Kitagawa T, Hokama Y, Suzuki T (1974) Protein Identification Resource (PIR). National Biomedical Research Foundation, Washington DC
Takahashi K, Gojobori T, Tanifuji M (1991) Two-color cytofluorometry and cellular properties of the urokinase receptor associated with a human metastatic carcinomatous cell line. Exp Cell Res 192:405–413
Takahata N, Kimura M (1981) A model of evolutionary base substitutions and its application with special reference to rapid change of pseudogenes. Genetics 98:641–657
Tanzi RE, McClatchey AI, Lamperti ED, Villa-Komaroff L, Gusella JF, Neve RL (1988) Protease inhibitor domain encoded by an amyloid protein precursor mRNA associated with Alzheimer's disease. Nature 33:1528–1530
Tschesche H, Dieti T (1975) The amino-acid sequence of isoinhibitor K from snails (Helix pomatia). Eur J Biochem 58: 439–451
Yamada T, Sasaki H, Dohura K, Goto I, Sakaki Y (1989) Structure and expression of alternatively-spliced forms of mRNA for the mouse homolog of Alzheimer's disease amyloid beta precursor. Biochem Biophys Res Commun 15:8906–8912
Wachter E, Deppner K, Hochstrasser K, Lempart K, Geiger R (1980) Protein Identification Resource (PIR). National Biomedical Research Foundation, Washington DC
Wun TC, Kretzmer KK, Girard TJ, Miletich JP, Broze GJ (1988) Cloning and characterization of a cDNA coding for the lipoprotein associated coagulation inhibitor shows that it consists of three tandem Kunitz-type inhibitory domains. J Biol Chem 263:6001–6004
Wunderer G, Machleidt W, Fritz H (1981) The broad-specificity proteinase inhibitor 5 II from the sea anemone Anemonia sulcata. Methods Enzymol 80:816–820
Author information
Authors and Affiliations
Additional information
Offprint requests to: T. Gojobori
Rights and permissions
About this article
Cite this article
Ikeo, K., Takahashi, K. & Gojobori, T. Evolutionary origin of a Kunitz-type trypsin inhibitor domain inserted in the amyloid β precursor protein of Alzheimer's disease. J Mol Evol 34, 536–543 (1992). https://doi.org/10.1007/BF00160466
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00160466