Abstract
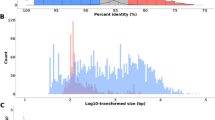
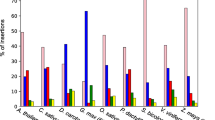
A surprisingly large number of plant nuclear DNA sequences inferred to be remnants of chloroplast and mitochondrial DNA migration events were detected through computer-assisted database searches. Nineteen independent organellar DNA insertions, with a median size of 117 by (range of 38 to >785 bp), occur in the proximity of 15 nuclear genes. One fragment appears to have been passed through a RNA intermediate, based on the presence of an edited version of the mitochondrial gene in the nucleus. Tandemly arranged fragments from disparate regions of organellar genomes and from different organellar genomes indicate that the fragments joined together from an intracellular pool of RNA and/or DNA before they integrated into the nuclear genome. Comparisons of integrated sequences to genes lacking the insertions, as well as the occurrence of coligated fragments, support a model of random integration by end joining. All transferred sequences were found in noncoding regions, but the positioning of organellar-derived DNA in introns, as well as regions 5′ and 3′ to nuclear genes, suggests that the random integration of organellar DNA has the potential to influence gene expression patterns. A semiquantitative estimate was performed on the amount of organellar DNA being transferred and assimilated into the nucleus. Based on this database survey, we estimate that 3–7% of the plant nuclear genomic sequence files contain organellar-derived DNA. The timing and the magnitude of genetic flux to the nuclear genome suggest that random integration is a substantial and ongoing process for creating sequence variation.
Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ayliffe MA, Timmis JN (1992a) Plastid DNA sequence homologies in the tobacco nuclear genome. Mol Gen Genet 236:105–112
Ayliffe MA, Timmis JN (1992b) Tobacco nuclear DNA contains long tracts of homology to chloroplast DNA. Theor Appl Genet 85:229–238
Baldauf SL, Palmer JD (1990) Evolutionary transfer of the chloroplast tufA gene to the nucleus. Nature 344:262–265
Bernatzky R, Mau S-L, Clarke AE (1989) A nuclear sequence associated with self-incompatibility in Nicotiana alata has homology with mitochondrial DNA. Theor Appl Genet 77:320–324
Bureau TE, Wessler SR (1994) Mobile inverted repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc Natl Acad Sci USA 91:1411–1415
Cheung WY, Scott NS (1989) A contiguous sequence in spinach nuclear DNA is homologous to three separated sequences in chloroplast DNA. Theor Appl Genet 77:625–633
Cushman JC, Bohnert HJ (1992) Salt stress alters A/T-rich DNA-binding factor interactions within the phosphoenolpyruvate carboxylase promoter from Mesembryanthemum crystallinum. Plant Mol Biol 20:411–424
Dangl JL, Hahlbrock K, Schell J (1989) Regulation and structure of chalcone synthase genes. Cell culture and somatic cell genetics of plants, vol 6: Genetics of plants. Academic Press, San Diego
Devereaux J, Haeberli P, Smithies O (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res 12: 387–395
Farrelly F, Butow RA (1983) Mitochondrial genes in the yeast nuclear genome. Nature 301:296–301
Fukuchi M, Shikanai T, Kossykh VG, Yamada Y (1991) Analysis of nuclear sequences homologous to the B4 plasmid-like DNA of rice mitochondria: evidence for sequence transfer from mitochondria to nuclei. Curr Genet 20:487–494
Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD (1991) Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J 10:3073–3078
Gellissen G, Bradfield JY, White BN, Wyatt GR (1983) Mitochondrial DNA sequences in the nuclear genome of a locust. Nature 301: 631–634
Gellissen G, Michaelis G (1987) Gene transfer: mitochondria to nucleus. Ann N Y Acad Sci 503:391–401
Gray MW (1992) The endosymbiotic hypothesis revisited. Int Rev Cytol 141:233–357
Gray MW, Covello PS (1993) RNA editing in plant mitochondria and chloroplasts. FASEB J 7:64–71
Grohmann L, Brennicke A, Schuster W (1992) The mitochondrial gene encoding ribosomal protein S12 has been translocated to the nuclear genome in Oenothera. Nucleic Acids Res 20:5641–5646
Hadler HI, Dimitrijevic B, Mahalingam R (1983) Mitochondrial DNA and nuclear DNA from normal rat liver have a common sequence. Proc Natl Acad Sci USA 80:6495–6499
Hua S, Dube SK, Kung S (1993) Sequence similarity in nuclear and mitochondrial gene regions in plants. J Plant Biochem Biotech 2:71–73
Knoop V, Brennicke A (1991) A mitochondrial intron sequence in the 5′-flanking region of a plant nuclear lectin gene. Curr Genet 20: 423–425
Knoop V, Brennicke A (1994) Promiscuous mitochondrial group II intron sequences in plant nuclear genomes. J Mol Evol 39:144–150
Koch I, Hofschneider PH, Koshy R (1989) Expression of a hepatitis B virus transcript containing fused mitochondrial-like domains in human heptoma cells. Virology 170:591–594
Koes RE, Spelt CE, van Den Elzen PJM, Mol JNM (1989) Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene 81:245–257
Margulis L (1993) Symbiosis in cell evolution: microbial communities in the Archeon and Protorozoic eons. Freeman, New York
Nakazono M, Hirai A (1993) Identification of the entire set of transferred chloroplast DNA sequences in the mitochondrial genome of rice. Mol Gen Genet 236:341–346
Nugent JM, Palmer JD (1991) RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell 66:473–481
Okubara PA, Williams SA, Doxsee RA, Tobin EM (1993) Analysis of genes negatively regulated by phytochrome action in Lemna gibba and identification of a promoter region required for phytochrome responsiveness. Plant Physiol 101:915–924
Olmstead RG, Michaels HJ, Scott KM, Palmer JD (1992) Monophyly of the Asteridae and identification of their major lineages inferred from DNA sequences of rbcL. Ann Missouri Botanical Gardens 79:249–265
Ossario PN, Sibley DL, Boothroyd JC (1991) Mitochondrial-like DNA sequences flanked by direct and inverted repeats in the nuclear genome of Toxoplasma gondii. J Mol Biol 22:525–536
Ott RW, Chua NH (1990) Enhancer sequences from Arabidopsis thaliana obtained by library transformation of Nicotiana tabacum. Mol Gen Genet 223:169–179
Palmer JD (1991) Plastid chromosomes: structure and evolution. In: Bogorad L, Vasil IK (ed) Cell culture and somatic cell genetics of plants, vol 7A: The molecular biology of plastids. Academic Press, San Diego, pp 5–53
Pichersky E (1990) Nomad DNA—a model for movement and duplication of DNA sequences in plant genomes. Plant Mol Biol 15: 437–448
Pichersky E, Logsdon Jr JM, McGrath JM, Stasys RA (1991) Fragments of plastid DNA in the nuclear genome of tomato: prevalence, chromosomal location, and possible mechanism of integration. Mol Gen Genet 225:453–458
Pichersky E, Tanksley SD (1988) Chloroplast DNA sequences integrated into an intron of a tomato nuclear gene. Mol Gen Genet 215:65–68
Roth D, Wilson (1988) Illegitimate recombination in mammalian cells. In: Kucherlapati R, Smith G (eds) Genetic recombination. American Society for Microbiology, Washington, DC
Schuster W, Brennicke A (1987) Plastid, nuclear and reverse transcriptase sequences in the mitochondrial genome of Oenothera: is genetic information transferred between organelles via RNA? EMBO J 6:2857–2863
Shay JW, Baba T, Zhan Q, Kamimura N, Cuthbert JA (1991) HeLaTG cells have mitochondrial DNA inserted into the c-myc oncogene. Oncogene 6:1869–1874
Stern DB, Lonsdale DM (1982) Mitochondrial and chloroplast genomes of maize and have a 12-kilobase DNA sequence in common. Nature 299:698–702
Sun C-W, Callis J (1993) Recent stable insertion of mitochondrial DNA into an Arabidopsis polyubiquitin gene by nonhomologous recombination. Plant Cell 5:97–107
Swofford DL (1993) Phylogenetic analysis using parsimony (PAUP), version 3.1.1. Illinois Natural History Survey, Champaign, IL
Thorsness PE, Fox TD (1993) Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondrial to the nucleus. Genetics 134:21–28
Thorsness PE, White KH, Fox TD (1993) Inactivation of YME1, a member of the ftsH-SECl8-PAST-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol 13:5418–5426
Timmis JN, Scott NS (1983) Sequence homology between spinach nuclear and chloroplast genomes. Nature 305:65–67
van den Boogaart P, Samallo J, Agsterbibe E (1982) Similar genes for a mitochondrial ATPase subunit in the nuclear and mitochondrial genomes of Neurospora crassa. Nature 298:187–189
Wintz H, Hanson M (1991) A termination codon is created by RNA editing in the petunia mitochondrial atp9 gene transcript. Curr Genet 19:61–64
Wolfe KH, Sharp PM, Li W-H (1989) Rates of synonymous substitution in plant nuclear genes. J Mol Evol 29:208–211
Zullo S, Leang CS, Slighton JL, Hadler HI, Eisenstadt JM (1991) Mitochondrial D-loop sequences are integrated in the rat nuclear genome. J Mol Biol 221:1223–1235
Author information
Authors and Affiliations
Additional information
Correspondence to: J.L. Blanchard
Rights and permissions
About this article
Cite this article
Blanchard, J.L., Schmidt, G.W. Pervasive migration of organellar DNA to the nucleus in plants. J Mol Evol 41, 397–406 (1995). https://doi.org/10.1007/BF00160310
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00160310