Abstract
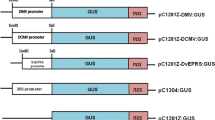
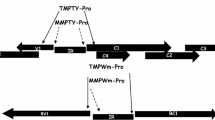
The cassava vein mosaic virus (CVMV) is a double stranded DNA virus which infects cassava plants (Manihot esculenta Crantz) and has been characterized as a plant pararetrovirus belonging to the caulimovirus subgroup. Two DNA fragments, CVP1 of 388 nucleotides from position -368 to +20 and CVP2 of 511 nucleotides from position -443 to +72, were isolated from the viral genome and fused to theuidA reporter gene to test promoter expression. The transcription start site of the viral promoter was determined using RNA isolated from transgenic plants containing the CVMV promoter:uidA fusion gene. Both promoter fragments were able to cause high levels of gene expression in protoplasts isolated from cassava and tobacco cell suspensions. The expression pattern of the CVMV promoters was analyzed in transgenic tobacco and rice plants, and revealed that the GUS staining pattern was similar for each construct and in both plants. The two promoter fragments were active in all plant organs tested and in a variety of cell types, suggesting a near constitutive pattern of expression. In both tobacco and rice plants, GUS activity was highest in vascular elements, in leaf mesophyll cells, and in root tips.
Similar content being viewed by others
References
Anza OA: Replication and mapping of caulimoviruses. PhD thesis, University of California, Davis, CA (1982).
Bao Y, Hull R: Mapping the 5′-terminus of rice tungro bacilliform viral genomic RNA. Virology 197: 445–448 (1993).
Battraw M, Hall T: Histochemical analysis of CaMV promoter-β-glucuronidase gene expression in transgenic rice plants. Plant Mol Biol 15: 527–538 (1990).
Benfey PN, Chua NH: The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue specific expression patterns. EMBO J 8: 2195–2202 (1989).
Benfey PN, Chua NH: The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250: 959–966 (1990).
Benfey PN, Ren L, Chua NH: Combinatorial and synergistic properties of CaMV 35S enhancer subdomains. EMBO J 9: 1685–1696 (1990).
Benfey PN, Ren L, Chua NH: Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J 9: 1677–1684 (1990).
Bhattacharyya-Pakrasi M, Peng J, Elmer JS, Laco G, Shen P, Kaniewska MB, Kononowicz H, Wen F, Hodges TK, Beachy RN: Specificity of a promoter from the rice tungro bacilliform virus for expression in phloem tissues. Plant J 4: 71–79 (1993).
Bradford MM: a rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye-binding. Anal. Biochem 72: 248–254 (1976).
Calvert L, Ospina M, Shepherd RJ: Characterization of cassava vein mosaic virus: a distinct plant pararetrovirus. J Gen Virol 76: 1271–1276 (1995).
Chen G, Müller M, Potrykus I, Hohn T, Fütter J: Rice tungro bacilliform virus: transcription and translation in protoplasts. Virology 204: 91–100 (1994).
Covey S, Hull R: Genetic engineering with double-stranded DNA viruses. In: Wilson TMA, Davies JWG (eds) Genetic Engineering with Plant Viruses, pp. 217–221. CRC Press, Boca Raton, FL (1992).
Donald G, Cashmore A: Mutation of either G box or I box sequences profoundly affects expression from theArabidopsis rbcs-1A promoter. EMBO J 9: 1717–1726 (1990).
Dowson J, Ashurst JL, Mathias SL, Watts JW, Wilson MA, Dixon RA: Plant viral leaders influence expression of a reporter gene in tobacco. Plant Mol Biol 23: 97–109 (1993).
Fang RX, Nagy F, Sivasubramaniam S, Chua NH: Multiplecis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell 1: 141–150 (1989).
Fromm H, Katagiri F, Chua NH: An octopine synthase enhancer element directs tissue-specific expression and binds ASF-1, a factor from tobacco nuclear extracts. Plant Cell 1: 977–984 (1989).
Fütterer J, Gordon K, Sanfaçon H, Bonneville J, Hohn T: Positive and negative control of translation by the leader sequence of cauliflower mosaic virus pregenomic 35S RNA. EMBO J 9: 1697–1707 (1990).
Gardner RC, Howarth AJ, Hahn P, Brown-Luedi M, Shepherd RJ, Messing J: The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing. Nucl Acids Res 9: 2871–2887 (1981).
Guilley H, Dudley RK, Jonard G, Balaszs E, Richards KE: Transcription of cauliflower mosaic virus DNA: detection of promoter sequences, and characterization of transcripts. Cell 30: 763–773 (1982).
Hay JM, Matthew CJ, Blakebrough M, Dasgupta I, Davies JW, Hull R: An analysis of the sequence of an infectious clone of rice tungro bacilliform virus, a plant pararetrovirus. Nucl Acids Res 19: 2615–2621 (1991).
Hohn T, Hohn B, Pfeiffer P: Reverse transcription in CaMV. Trends Biochem Sci 10: 205–209 (1985).
Horsch RB, Fry J, Hoffmann N, Neidermeyer J, Rogers SG, Fraley RT: Leaf disc transformation. In Gelvin SB, Schilperoort RA (eds) Plant Molecular Biology Manual, pp. A5/1-A5/9. Kluwer Academic Publishers, Dordrecht, Netherlands (1988).
Hull R: Caulimovirus group. CMI/ABB, Description of plant viruses no. 229 (1984).
Jefferson RA, Kavanagh TA, Bevan MW: Gus fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 (1987).
Katagiri F, Seipel K, Chua NH: Identification of a novel dimer stabilization region in a plant bZIP transcription activator. Mol Cell Biol 12: 4809–4816 (1992).
Kay R, Chan A, Daly M, McPherson J: Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236: 1299–1302 (1987).
Kikkawa H, Nagata T, Matsui C, Takebe I: Infection of protoplasts from tobacco suspension culture by tobacco mosaic virus. J Gen Virol 63: 457–467 (1982).
Kitajima EW, Costa AS: Particulas esferoidais associadas ao virus do mosaico das nervuras da mandioca. Bragantia 25: 211–221 (1966).
Lam E, Benfey PN, Gilmartin PM, Fang RX, Chua NH: Sitespecific mutations alterin vitro factor binding and change promoter expression pattern in transgenic plants. Proc Natl Acad Sci US`A 86: 7890–7894 (1989).
Leckie F, Devoto A, Delorenzo G: Normalization of GUS by luciferase activity from the same cell extract reduces transformation variability. Bio Techniques 17: 54–57 (1994).
Li L, Qu R, deKochko A, Fauquet C, Beachy R: An improved rice transformation system using the biolistic method. Plant Cell Rep 12: 250–255 (1993).
Lockart BEL: Evidence for a double-stranded circular DNA genome in a second group of plant viruses. Phytopathology 80: 545–550 (1990).
Medberry SL, Lockhart BEL, Olszewski NE: Properties ofCommelina yellow mottle virus's complete DNA sequence, genomic discontinuities and transcript suggest that it is a pararetrovirus. Nucl Acids Res 18: 5505–5513 (1990).
Medberry SL, Lockhart BEL, Olszewski NE: TheCommelina yellow mottle virus promoter is a strong promoter in vascular and reproductive tissues. Plant Cell 4: 185–192 (1992).
Medberry SL, Olszewski NE: Identification of cis-elements involved in Commelina yellow mottle virus promoter activity. Plant J: 619–626 (1993).
Murashige T, Skoog F: A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 (1962).
Odell JT, Nagy F, Chua NH: Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313: 810–812 (1985).
Ondek B, Shepard A, Herr W: Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J 6: 1017–1025 (1987).
Ow DW, Jacobs JD, Howell SH: Functional regions of the cauliflower mosaic virus 35S RNA promoter determined by use of the firefly luciferase gene as a reporter of promoter activity. Proc Natl Acad Sci USA 84: 4870–4874 (1987).
Ow DW, Wood HV, DeLuca M, DeWet JR, Helinski DR, Howell SH: Transient and stable expression of the firefly luciferase gene in plant cells and transgenic plants. Science 234: 856–859 (1986).
Prescot A, Martin C: A rapid method for the quantitative assessment of levels of specific mRNAs in plants. Plant Mol Biol Rep 4: 219–224 (1987).
Qu R, Bhattacharyya M, Laco G, DeKochko A, Subba Rao BL, Kaniewska MB, Elmer S, Rochester DE, Smith CE, Beachy RN: Characterization of the genome of rice tungro bacilliform virus: comparison withCommelina yellow mottle virus and caulimoviruses. Virology 185: 354–364 (1991).
Richins RD, Scholthof HB, Shepherd RJ: Sequence of figwort mosaic virus DNA (caulimovirus group). Nucl Acids Res 15: 8451–8466 (1987).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Sanfaçon H: Analysis of figwort mosaic virus (plant pararetrovirus) polyadenylation signal. Virology 198: 39–49 (1994).
Sanger M, Daubert S, Goodman RM: Characteristics of a strong promoter from figwort mosaic virus: comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol Biol 14: 433–443 (1990).
Singh A, Kao TH, Lin JJ. Transformation ofAgrobacterium tumefaciens with T DNA vector using high-voltage electroporation. Focus 15: 84–87 (1993).
Taylor NJ, Edwards M, Klernan RJ, Davey C, Blakesley D, Henshaw GG. Development of friable embryogenic callers and embryogenic suspension cultures in cassava (Manihot esculenta) (rantz). Nature Biotechnology 14: 726–730 (1996).
Terada R, Shimamoto K: Expression of CaMV35S-GUS gene in transgenic rice plants. Mol Gen Genet 220: 389–392 (1990).
Yanagisawa S, Katsura I: MNFI, a lcaf tissue-specific DNA-binding protein of maize, interacts with the cauliflower mosaic virus 35S promoter as well as the C4 photosynthetic phosphoenolpyruvate carboxylase gene promoter. Plant Mol Biol 19: 545–553 (1992).
Yin Y, Beachy R: The regulatory regions of the rice tungro bacilliform virus promoter and interacting nuclear factor in rice (Oryza sativa L.). Plant J 7: 969–980 (1995).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Verdaguer, B., de Kochko, A., Beachy, R.N. et al. Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol Biol 31, 1129–1139 (1996). https://doi.org/10.1007/BF00040830
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00040830