Abstract
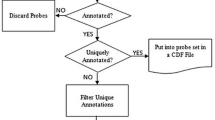
We have compared Affymetrix and Bioconductor annotations for the MOE430A (mouse) GeneChip® array. The mappings of probe sets to LocusLink identifiers (LocusIDs) were found to be dynamic, with many changes between successive releases of annotation for both Affymetrix and Bioconductor. There are 49 probe sets that are assigned to one LocusID by Affymetrix and to a different LocusID by Bioconductor from mid-2004 onwards. For virtually all of these examples, the Affymetrix annotation was found to be the one that is in agreement with the current gene prediction.
Reference sequence (RefSeq) identifiers are considered to be the gold standard of annotations. However, we could not use these identifiers to discriminate between the accuracy of Bioconductor and Affymetrix because not all of the probes map to the RefSeq transcript to which the probe set is assigned. Moreover, in some cases, probes align to regions downstream of the 3′ end of a RefSeq transcript.
Adjacent genes were found to be a major cause of discrepancies between the Bioconductor and Affymetrix assignments. Case studies of several probe sets indicated that incorrect assignments are caused by the UniGene cluster assignments of expressed sequence tags representing the probe sets, and by errors in GenBank® sequences.
Our results indicate that there are a number of errors remaining in the annotation sources used by the microarray community.













Similar content being viewed by others
References
Affymetrix, Inc. GeneChip® Custom Express™ array design guide: part no. 700506 rev. 4 [online]. Available from URL: http://www.affymetrix.com/support/technical/other/custom_design_manual.pdf [Accessed 2006 Sep 27]
Affymetrix, Inc. Array design and performance of the GeneChip® mouse expression set 430: technical note, part no. 701405 Rev. 1 [online]. Available from URL: http://www.affymetrix.com/support/technical/technotes/mouse430_technote.pdf [Accessed 2006 Oct 9]
GenBank® [online]. Available from URL: http://www.ncbi.nlm.nih.gov/Genbank/index.html [Accessed 2006 Sep 26]
NCBI expressed sequence tags database [online]. Available from URL: http://www.ncbi.nlm.nih.gov/dbEST/ [Accessed 2006 Sep 26]
NCBI reference sequences [online]. Available from URL: http://www.ncbi.nlm.nih.gov/RefSeq [Accessed 2006 Sep 26]
NCBI UniGene [online]. Available from URL: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?.db=unigene [Accessed 2006 Sep 26]
Affymetrix, Inc. Array design for the GeneChip® human genome U133 set: technical note, part no. 701133 rev. 1 [online]. Available from URL: http://www.affymetrix.com/support/technical/technotes/hgu133_design_technote.pdf [Accessed 2006 Oct 9]
Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res 2001; 29: 137–40
Maglott D, Ostell J, Pruitt KD, et al. Entrez Gene: gene-centred information at NCBI. Nucleic Acids Res 2005; 33 (database issue): D54–8
Pontius JU, Wagner L, Schuler GD. UniGene: a unified view of the transcriptome. In: The NCBI handbook, 2003 [online]. Available from URL: http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=handbook.chapter.ch21 [Accessed 2006 Oct 10]
Liu G, Loraine AE, Shigeta R, et al. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res 2003; 31: 82–6
Affymetrix, Inc. Transcript assignment for Affymetrix GeneChip® probe arrays [online]. Available from URL: http://www.affymetrix.com/support/technical/whitepapers/netaffxannot_whitepaper.pdf [Accessed 2006 Sep 27]
Kent WJ. BLAT: the BLAST-like alignment tool. Genome Res 2002; 12: 656–64
Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5(10): R80
Zhang J. Basic functions of AnnBuilder [online]. Available from URL: http://www.bioconductor.org/repository/devel/vignette/AnnBuilder.pdf [Accessed 2006 Sep 27]
Zhang, J. How to use AnnBuilder [online]. Available from URL: http://www.bioconductor.org/repository/devel/vignette/ABPrimer.pdf [Accessed 2006 Sep 27]
Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 2002; 420: 520–62
UCSC Genome Bioinformatics [online]. Available from URL: http://genome.ucsc.edu [Accessed 2006 Sep 26]
Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res 2002; 12: 996–1006
Karolchik D, Baertsch R, Diekhans M, et al. The UCSC genome browser database. Nucleic Acids Res 2003; 31: 51–4
Ensembl [online]. Available from URL: http://www.ensembl.org/index.html [Accessed 2006 Sep 26]
Hubbard T, Andrews D, Caccamo M, et al. Ensembl 2005. Nucleic Acids Res 2005; 33 (database issue): D447–53
Interface AffyProbe [online]. Available from URL: http://www.ensembl.org/info/software/java/api/org/ensembl/datamodel/AffyProbe.html [Accessed 2006 Oct 11]
Affymetrix. Mouse expression set 430 [online]. Available from URL: http://www.affymetrix.com/support/technical/byproduct.affx?product=moe430 [Accessed 2006 Oct 10]
Affymetrix. Rat expression set 230 [online]. Available from URL: http://www.affymetrix.com/support/technical/byproduct.affx?product=rae230 [Accessed 2006 Oct 10]
NetAffx analysis center [online]. Available from URL: http://www.affymetrix.com/analysis/index.affx [Accessed 2006 Oct 10]
Ihaka R, Gentleman R. R: a language for data analysis and graphics. Journal of Computational and Graphical Statistics 1996; 5: 299–314
NCBI Map Viewer [online]. Available from URL: http://www.ncbi.nlm.nih.gov/mapview [Accessed 2006 Sep 26]
NCBI UniGene FAQ [online]. Available from URL: http://www.ncbi.nlm.nih.gov/UniGene/FAQ.shtml [Accessed 2006 Sep 27]
Ensembl. Microarray probeset mapping [online]. Available from URL: http://www.ensembl.org/info/data/docs/microarray_probe_set_mapping.html [Accessed 2006 Oct 16]
Perez-Iratxeta C, Andrade MA. Inconsistencies over time in 5% of NetAffx probeto-gene annotations. BMC Bioinformatics 2005; 6: 183
Harbig J, Sprinkle R, Enkemann SA. A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array. Nucleic Acids Res 2005; 33: e31
Acknowledgements
This work was partially funded by the Biotechnology and Biological Sciences Research Council, UK. The authors have no conflicts of interest that are directly relevant to the contents of this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Stalteri, M., Harrison, A. Comparisons of Annotation Predictions for Affymetrix GeneChips®. Appl-Bioinformatics 5, 237–248 (2006). https://doi.org/10.2165/00822942-200605040-00006
Published:
Issue Date:
DOI: https://doi.org/10.2165/00822942-200605040-00006