Abstract
Background
In Nepal, cases of Cholera occur annually either as sporadic or as outbreaks claiming the lives of many in rural areas. The present study is a laboratory based surveillance which aims to analyze the changing epidemiology and antimicrobial susceptibility trend of V. cholerae strains isolated or referred to National Public Health Laboratory (NPHL) over a period of 11 years (2006–2016).
Methods
Specimens of fresh stool /rectal swab either received at sentinel sites or NPHL were processed following standard microbiological techniques. Suspected colonies on selective medium were identified using routine biochemical tests and confirmed by serotyping. Antimicrobial susceptibility testing was performed following Kirby Baeur disc diffusion method.
Results
Of the 836 confirmed isolates, 87% (728/836) were V.cholerae O1 Ogawa,12% (103/836) were V.cholerae O1 Inaba and only 6 isolates were V.cholerae O1 Hikojima. In 2006 all the Vibrio isolates were of Inaba serotype, followed by all 3 serotypes during 2007.During 2008–2014 only Ogawa serotype was isolated while few cases of Inaba again surfaced in 2015. Resistance to ampicillin decreased from 93% in 2006 to 18% by 2010 and again raised to 100% by 2016.Cotrimoxazole resistance remained at constant range (77–100%).Nalidixic acid resistance was 100% since 2006.Ciprofloxacin and tetracycline resistance emerged in 2007, reached a peak during 2010–2012 and declined to 0 by 2016.Susceptibility to Furazolidone has re-emerged.63.6% of the isolates were Multi drug resistant.
Conclusion
With changing epidemiology and antibiogram of V.cholerae in Nepal, the present study reflects the importance of continuous monitoring, which could be used by policy makers and health professionals for better management of outbreaks. Decline in tetracycline and ciprofloxacin resistance along with emerging sensitivity to furazolidone shows that these drugs could make an effective comeback in future.
Similar content being viewed by others
Background
Cholera is an acute diarrheal illness caused by the bacterium Vibrio cholerae [1]. Until now, seven distinct pandemics of cholera have been recorded [2]. The earlier pandemics were caused by the classical biotype of V. cholerae O1, whereas the 7th ongoing pandemic is caused by serogroup O1 El Tor and has been the most extensive in geographical spread and duration [3]. Outbreak of epidemic cholera caused by a non-O1 strain of V. cholerae designated V. cholerae 0139 has also been reported from various parts of Bangladesh, India and other countries [4]. Globally, there are an estimated 3–5 million cholera cases and 100,000–120,000 deaths every year. Commonly, lack of prompt, proper treatment leads to shock within 6–12 h followed by death occurring between18 h and several days [5].
Over the past few years, V. cholerae O1 biotype El Tor causing Asiatic cholera has shown remarkable changes in its phenotypic and genetic characteristics [6]. The most recent development in the evolution of global cholera has been the emergence and spread of a new variant ET, or altered ET, in Bangladesh that carries ctxB of the classical (CL) biotype (ctxBCL) [7]. According to recent reports, altered ET strains have been spreading globally [8,9,10,11,12,13,14,15] causing more severe disease [16].
In Nepal, cholera is endemic. Every year, cases of cholera occur sporadically or as outbreaks. Diarrhea and gastroenteritis (including cholera and dysentery) comes in second position among top ten causes of inpatient morbidity and top ten reasons for OPD consultation. in Nepal (DoHS, 2017). It has also been included among one of the six outbreak potential diseases and are reported immediately through the Early Warning Reporting System (EWARS) to the National Epidemiology and Disease Control division (EDCD, 2017). In a decade, six episodes of laboratory confirmed cholera outbreak has been recorded by NPHL.
Methods
-
A.
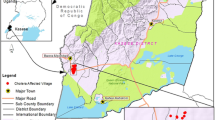
Surveillance sites: There are 22 sentinel sites participating in laboratory based national antimicrobial resistance (AMR) surveillance. V.cholerae was isolated at 11 participating laboratories (Fig. 1)
-
B.
Source of Vibrio:
Suspected stool samples received at surveillance sites and samples from suspected outbreaks received at the reference laboratory were processed following standard microbiological procedures as mentioned below. Presumptive isolates from any source was confirmed, serotyped and stored at the reference laboratory.
-
C.
Laboratory Methods:
-
1.
Sample processing:
Fresh stool/rectal swab samples were parallelly added to 5 ml of alkaline peptone water (APW) and streaked onto MacConkey agar and Thiosulphate Citrate Bile Salt Sucrose (TCBS) agar. Thus inoculated APW was incubated at 35–37 °C for 6–8 h for enrichment and the plates were incubated at 35–37 °C for 18–24 h.
After enrichment, a loopful of APW was inoculated onto MacConkey agar and TCBS agar and then incubated at 35–37 °C for 18–24 h. Suspected colonies (i.e non-lactose fermenting on MacConkey agar or golden yellow colonies on TCBS) were identified by Gram stain, oxidase test, string test and inoculation on biochemical medium viz. Triple Sugar Iron agar, SIM medium, Urease agar, Citrate agar and MR-VP broth.
-
2.
Serotyping: All isolates of Vibrio were confirmed serologically by slide agglutination using serogroup specific O1 polyvalent and O139 antisera and monospecific antisera for Ogawa & Inaba strains from Denka Seiken Company Limited, Tokyo, Japan. Strains showing visible agglutination were considered positive.
-
3.
Biotyping: Vibrio isolates were biotyped using Vogues-proskaeur reaction and sensitivity to Polymyxin B (50 units) discs.

Flowchart for sample processing
-
D.
Antibiotic susceptibility test was performed for all the identified Vibrio isolates by standard Kirby Bauer’s Disc Diffusion technique following updated CLSI guidelines [17]. The antibiotics tested were ampicillin (AMP-10mcg), cotrimoxazole (COT-25mcg), nalidixic acid (NA- 30mcg), ciprofloxacin (CIP-5mcg), tetracycline (T-30mcg), furazolidone (FR-100 units) and ceftriaxone (CRO-5 mcg). Strains of Escherichia coli ATCC 25922 were used as control.
-
E.
Statistical methods: Data obtained were analyzed using SPSS software for windows version 18.Comparison of data with respect to Vibrio, sex, and age-groups were performed by Chi- square. P < 0.05 was considered to be statistically significant. Duplicate samples were counted as one during data entry.
Results
Nepal reported 6 outbreaks in 18 districts (of 77), on 6 out of 7 provinces (Fig. 1), from 2006 to 2016. A total of 836 V.cholerae isolates were reported in a period of 11 years (2006–2016) from 11 participating laboratories including NPHL. The highest number was reported in 2007(Fig. 1). Among the participating laboratories, 42% were reported from Patan hospital, followed by NPHL and Sukraraj tropical and Infectious Disease Hospital (Fig. 2).
All the isolates verified at NPHL were of O1 serogroup and Eltor biotype. Of the total 87% (724) were V.cholerae O1 Ogawa, 12% (103) were V.cholerae O1 Inaba and only 1% [6] isolates were of V.cholerae O1 Hikojima serotype. However the prevalent serotype kept changing over the years. In 2006, V. cholerae O1 Inaba was predominant, by 2007 all serotypes V. cholerae O1 Eltor Ogawa, Inaba & Hikojima were observed. During 2008–2016, V. cholerae O1 Ogawa predominated again with one case each of V. cholerae O1 Inaba in recent years (Table 1).
Cholera cases peaked in monsoon season (June to august). Overall, infection was higher in males as compared to females but no significant difference was observed. Among both the gender, infection was predominant in 21–30 years age group (Fig. 3).
Rapid fluctuation in antimicrobial susceptibility trend among Vibrio cholerae was observed over the years and is demonstrated in Fig. 4.Resistance to ampicillin decreased from 93% in 2006 to 18% by 2010 and again rose to 100% by 2016. Cotrimoxazole resistance fluctuated between 76 and 100%. All the isolates were sensitive to ciprofloxacin till 2006; resistance emerged in 2007, peaked during 2012 and declined to 0 by 2016. After several years in which V. cholerae was uniformly susceptible to tetracycline, a sudden upsurge in tetracycline resistance was noted, from 0% in 2006 to 40% in 2007 before decreasing to 8% in 2009. A significant decline in furazolidone resistant isolates was observed in this study period (100% in 2006 to 5% in 2016). All the isolates were sensitive to ceftriaxone; however since 2013, isolates showing partial susceptibility has also been reported. In 2016, one isolate was resistant to ceftriaxone.
Of the total, 61.9% isolates (518/836) were Multidrug resistant. Of them, 4.1% (21/518) were resistant to five classes of antibiotics, 34.5% (179/518) were resistant to four classes and 61% (357/532) were resistant to three classes of antibiotics tested. The most common resistotype was isolates exhibiting simultaneous resistance to ampicillin, cotrimoxazole and quinolones (Table 2).
Discussion
Cholera, its sudden and explosive onset in the form of an outbreak or epidemic, coupled with high mortality and morbidity rates, still persists as a major health problem in Nepal. The present article gives a review of the cholera scenario based on biotype, serotype and antimicrobial susceptibility in our area from 2006 to 2016.
All the V.cholerae isolated over 11 years was of O1 serogroup, no isolate of non-O1 serogroup has been reported from Nepal so far. However, outbreaks due to non-O1 serogroups have been reported from India and Bangladesh [18,19,20,21]. In our study, all the V.cholerae isolates received and verified at NPHL were of El Tor biotype, which is in accordance to other studies [22,23,24,25,26] but contrary to study made by Rojina et al., in 2012, which reports 22 V.cholerae isolated at Sukraraj tropical and Infectious disease hospital to be of Classical Biotype [27]. Since, the hospital was included in AMR surveillance since 2013 only, the isolates were reported but not verified at NPHL.
Over a period of 11 years, interesting changes in prevalent serotype of V.cholerae was observed. Studies made before 2005, revealed Ogawa as the predominant serotype whereas during 2006, Inaba predominated. By 2007, all three serotypes were found. After nearly 9 years, by 2015 and 2016, one case of Inaba was seen, which may be an indication that gradually there may again be a conversion. Periodic shifts in the incidence of Ogawa and Inaba serotypes in a given area is a common phenomenon and is thought to be a consequence of genetic reversal that occurs both in vitro and in vivo [28, 29]. Such observation has been reported from various national and international studies [18, 30,31,32].
Up to now, all the reported outbreak and sporadic cases were mostly concentrated in Central and Far-western part of the country; however, few cases were also reported from eastern part. The hill districts of the mid and far western development regions are particularly affected due to its remoteness, poor and difficult accessibility, inadequate public health facilities and poor water and sanitation conditions. In the central region the supply of safe drinking water is inadequate due to increase in urban population density due to which people are forced to drink water from any available source with or without prior treatment. Other factors like floods, landslides, open defecation practices, breakdown in sanitation and in the supply of safe water also contribute to frequent cases of cholera [33].
Overall infection was higher in males, of adult age group (21–30 yrs). Study conducted by Uthappa et al. also supports 12% attack rate in males, but higher incidence in children of 6–14 years age group. Studies made by Bhandari et al. and Gupta et al. in Nepal have reported 33.3% in 15–44 years age group and 48.8% in 20–39 years age group, similar to our study [33, 34].
Resistance patterns in V. cholerae are known to fluctuate rapidly because it cannot stably carry plasmids that confer resistance and because the organism naturally resides in aquatic environments devoid of selective pressure from antibiotics [35, 36]. Our study has also shown dramatic fluctuations in resistance to routinely used antibiotics. High resistance against cotrimoxazole, nalidixic acid and furazolidone (90–100%) was evident since 2006. Such high resistances to cotrimoxazole and nalidixic acid have also been reported from Mozambique [37, 38], Zambia [39], India [40,41,42] and Haiti [43]. Along the years, ampicillin resistance fluctuated. Some studies have shown 100% resistance [39, 44] while some has shown increasing resistance [45].
The reason behind the sudden upsurge and subsequent decline in ciprofloxacin and tetracycline resistance is unknown; however, the decline in tetracycline resistance has also been reported from other areas [40, 42, 43]. Studies made by Karlsson et al. [43] and Mala et al. [46] have reported susceptibility to tetracycline similar to our current scenario. Similarly, studies conducted at Mozambique [39], Bangladesh [47] also shows susceptibility to ciprofloxacin, whereas some other studies show resistance fluctuating between 2 and 39% [41].
One of the important finding in our study was re-emergence of sensitivity to furazolidone. During our study period, furazolidone resistance remained 80–100% (except in 2012), and abruptly in 2016, a significant decline in resistance to 6% was noted. Klontz et al., in his study, have stated that they stopped consistently testing for resistance to furazolidone due to high resistance levels [47]. Most of the previous national and international studies have reported 100% isolates resistant to furazolidone [23, 48]. Some other studies have shown increasing resistance from 21% in 2008 to 95% in 2010 [45]. Recently, only 1 Vibrio was resistant to ceftriaxone, which may be an indication that, in near future, ceftriaxone resistance may also emerge. So far, no study has reported ceftriaxone resistance in Vibrio.
In our context, the origin of MDR Vibrio strain is not documented, however, studies made before 2006, have reported MDR (defined it as simultaneous resistance to 2 different classes of antibiotics) cholera cases [25]. With the current definition of MDR (isolates resistant to one or more of 3 or more different classes, our result shows that 94% isolates were MDR in 2006 reaching to 100% by 2016 which is in concordance to results obtained by Shrestha et al. [49]. Since, cholera was well prevalent in Nepal before this period, MDR strains may have emerged presumably due to heavy and widespread use of antibiotics for therapy.
Conclusion
Since the emergence of antibiotic resistance may significantly influence patient care, continuous monitoring of epidemic strains is crucial. Molecular studies emphasizing on the Although few isolates were further characterized for ctxB gene, MLVA typing, the genes and mechanisms responsible for antibiotic resistance still remains an area for further exploration.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- Amp:
-
Ampicillin
- AMR:
-
Antimicrobial Resistance
- APW:
-
Alkaline Peptone Water
- Cot:
-
Cotrimoxazole
- CRO:
-
Ceftriaxone
- Ctx-B:
-
Cholera enterotoxin subunit B
- EDCD:
-
Epidemiology and Disease Control Division
- EWARS:
-
Early Warning Reporting System
- FR:
-
Furazolidone
- MDR:
-
Multidrug Resistant
- MLVA:
-
Multiple-Locus Variable number tandem repeat analysis
- MR-VP:
-
Methyl Red-Vogues Proskaeur
- NA:
-
Nalidixic Acid
- NPHL:
-
National Public Health Laboratory
- SIM:
-
Sulfur Indole Motility
- T:
-
Tetracycline
- TCBS:
-
Thiosulphate Citrate Bile Salt Sucrose
- TSI:
-
Triple Sugar iron
References
Kumar A, Oberoi A. Vibrio isolates from cases of acute diarrhea and their antibiotic susceptibility pattern in a tertiary care hospital of Punjab. CHRISMED Journal of Health and Research. 2014;1(4):254–7.
Pollitzer R, Swaroop S, Burrows W. Cholera. Monogr Ser World Health Organ. 1959;58(43):1001–19.
Shaban L, Ahmed S F, Materu S and Klena J D \, BMC Proc 2009, 3(Suppl 3):O9.
Calia KE, Murtah M, Ferrararo MJ, Calderwood SB. Comparison of Vibrio cholerae 0139 with V. cholerae 01 Classical and El Tor Biotypes. Infect Immun. 1994;62(4):1504–6.
World Health Organisation. Factsheet. 2012.
Nair GB, Faruque SM, Bhuiyan NA, Kamruzzaman M, Siddique AK, Sack DA. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol. 2002;40:3296–9.
Nair GB, Qadri F, Holmgren J, Svennerholm AM, Safa A, Bhuiyan NA, Ahmad QS, Faruque SM, Faruque ASG, Takeda Y, Sack DA. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol. 2006;44:4211–3.
Ansaruzzaman M, Bhuiyan NA, Nair GB, Sack DA, Lucas M, Deen JL, Ampuero J, Chaignat CL. The Mozambique cholera vaccine demonstration project coordination group. Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis. 2004;10:2057–9.
Safa A, Sultana J, Cam PD, Mwansa JC, Kong RYC. Vibrio cholerae O1hybrid El Tor strains, Asia and Africa. Emerg Infect Dis. 2008;14:987–8.
Nguyen BM, Lee JH, Cuong NT, Choi SY, Hien NT, Anh DD, Lee HR, Ansaruzzaman M, Endtz HP, Chun J, Lopez AL, Czerkinsky C, Clemens JD, Kim DW. Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J Clin Microbiol. 2009;47:1568–71.
Alam M, Nusrin S, Islam A, Bhuiyan NA, Rahim N, Delgado G, Morales R, Mendez JL, Navarro A, Gil AI, Watanabe H, Morita M, Nair GB, Cravioto A. Cholera between 1991 and 1997 in Mexico was associated with infection by classical, El Tor, and El Tor variants of Vibrio cholerae. J Clin Microbiol. 2010;48:3666–74.
Morita M, Ohnishi M, Arakawa E, Yamamoto S, Nair GB, Matsushita S, Yokoyama K, Kai A, Seto K, Watanabe H, Izumiya H. Emergence and genetic diversity of El Tor Vibrio cholerae O1 that possess classical biotype ctxB among travel associated cases of cholera in Japan. J Med Microbiol. 2010;59:708–12.
Okada K, Chantaroj S, Roobthaisong A, Hamada S, Sawanpanyalert P. A cholera outbreak of the Vibrio cholerae O1 El Tor variant carrying classical ctxB in northeastern Thailand in 2007. Am J Trop Med Hyg. 2010;82:875–8.
Goel AK, Jain M, Kumar P, Sarguna P, Bai M, Gosh N, Gopalan N. Molecular characterization reveals involvement of altered El Tor biotype Vibrio cholerae O1 strains in cholera outbreak at Hyderabad, India. J Microbiol. 2011;49:280–4.
Islam MS, Mahmud ZH, Ansaruzzaman M, Faruque SM, Talukder KA, Qadri F, Alam M, Islam S, Bardhan PK, Mazumder RN, Khan AI, Ahmed S, Iqbal A, Chitsatso O, Mudzori J, Patel S, Midzi SM, Charimari L, Endtz HP, Cravioto A. Phenotypic, genotypic and antibiotic sensitivity patterns of strains isolated from the cholera epidemic in Zimbabwe. J Clin Microbiol. 2011;49:2325–7.
Siddique AK, Nair GB, Alam M, Sack DA, Huq A, Nizam A, Longini IM, Qadri F, Faruque SM, Colwell RR, Ahmed S, Iqbal A, Bhuiyan NA, Sack RB. El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol Infect. 2010;138:347–52.
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100. Wayne: Clinical and Laboratory Standards Institute; 2018.
Narang P, Mendiratta DVS, Narang R. Changing pattern of Vibrio cholerae in Sevagram between 1990 and 2005. India J Med Microbiol. 2008;26:40–4.
Garg P, Nandy RK, Chaudhury P, Chowdhury NR, De K, Ramamurthy T. Emergence of Vibrio cholerae 01 biotype El Tor serotype Inaba from the prevailing 01 Ogawa serotype strains in India. J Clin Microbiol. 2000;38:4249–53.
Shimada T, Balakrish Nair G, Deb BC, Albert MJ, Sack RB. Outbreak of Vibrio cholerae non-O1 in India and Bangladesh. Lancet. 1993;341:1347 (Letter.).
Albert MJ, Siddique AK, Islam MS, Faruque AS, Ansaruzzaman M, Faruque SM, Sack RB. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341(8846):704.
Tamang MD, Sharma N, Makaju RK, Sharma AN, Koju R, Nepali N, Mishra SK. An outbreak of El Tor cholera in Kavre district, Nepal. Kathmandu Univ Med J. 2005;3:138–42.
Shah BK, Sharma S, Shakya G, Upadhyay BP. Multiple drug resistant Vibrio cholerae, Salmonella and Shigella from Nepalgunj cholera outbreak and different hospitals of Nepal. Nepalese J Biosciences. 2012;2:31–9.
Kansakar P, Baral P, Malla S, Ghimire GR. Animicrobial susceptibilities of enteric bacterial pathogens isolated in Kathmandu, Nepal, during 2002-2004. J Infect Dev Ctries. 2011;5:163–8.
Shrestha SD, Malla S, Adhikari BR, Shakya G, Basnyat SR, Sharma S. Antibiotic susceptibility patterns of Vibrio cholerae isolates. J Nepal Med Assoc. 2010;49:232–6.
Garg P, Sinha S, Chakraborty R, Bhattacharya SK, Nair GB, Ramamurthy T, Takeda Y. Emergence of fl uoroquinolone-resistant strains of Vibrio cholerae O1 biotype El Tor among hospitalized patients with cholera in Calcutta, India. Antimicrob Agents Chemother. 2001;45:1605–6.
Maharjan R, Shrestha D, Acharya J, Adhikari N, Rijal KR, Ghimire P, Shrestha UT. Change in biotype trend of Vibrio cholerae in Nepal. Med Micro J Microbiol. 2015;1(1):46–54.
Sack RB, Miller CE. Progressive changes of Vibrio serotypes in germ free mice infected with Vibrio cholerae. J Bacteriol. 1969;99:688–95.
Colwell RR, Huq A, Chowdhary MA, Brayton PR, Xu B. Serogroup conversion of Vibrio cholerae. Can J Microbiol. 1995;41:946–50.
Sarkar BL, Roy MK, Chakraborti AK, Niyogi SK. Distribution of phage type of Vibrio cholerae O1 biotype El tor in Indian scenario (1991–98). Indian J MedRes. 1999;109:204–7.
Shrestha SD, Malla S, Basnyat SR. Etiology of Diarrhoea with reference to multiple drug resistant enteric bacterial pathogens. Nepal J Sci Technol. 2008;9:131–8.
Malla S, Kansakar P, Ghimirey G. Cholera outbreak in Kathmandu valley in 2004: a review of National Public Health Laboratory findings. J Nepal Assoc Med Lab Sci. 2005;7:20–3.
Bhandari GP, Dixit SM, Ghimire U, Maskey MK. Outbreak investigation of diarrheal diseases in Jajarkot. J Nepal Health Res Council. 2009;7(15):66–8.
Gupta PK, Pant ND, Bhandari R, Shrestha P. Cholera outbreak caused by drug resistant Vibrio cholerae serogroup O1 biotype El Tor serotype Ogawa in Nepal; a cross-sectional study. Antimicrob Resist Infect Control. 2016;5:23.
Sack RB, Rahman M, Yunus M, Khan EH. Antimicrobial resistance in organisms causing diarrheal disease. Clin Infect Dis. 1997;24(suppl 1):S102–5.
David A, Sack CL, McLaughlin C, Suwanvanichki V. Antimicrobial resistance in shigellosis, cholera and campylobacteriosis. Geneva: World Health Organization; 2001.
Mandomando I, Espasa M, Valles X, Sacarlal J, Sigauque B, Ruiz J, et al. Antimicrobial resistance of Vibrio cholerae O1 serotype Ogawa isolated in Manhica District hospital, southern Mozambique. J Antimicrob Chemother. 2007;60(3):662–4.
Dengo-Baloi LC, Sema-Baltazar CA, Manhique LV, Chitio JE, Inguane DL, Langa JP. Antibiotics resistance in El Tor Vibrio cholerae 01 isolated during cholera outbreaks in Mozambique from 2012 to 2015. PLoS One. 2017;12(8):e0181496. https://doi.org/10.1371/journal.pone.0181496 e Collection 2017.
Mwansa JCL, Mwaba J, Lukwesa C, Bhuiyan NA, Ansaruzzaman M, Ramamurthy T, et al. Multiply antibiotic-resistant Vibrio cholerae O1 biotype El Tor strains emerge during cholera outbreaks in Zambia. Epidemiol Infect. 2007;135(05):847.
Garg P, Chakraborty S, Basu I, Datta S, Rajendran K, Bhattacharya T, et al. Expanding multiple antibiotic resistance among clinical strains of Vibrio cholerae isolated from 1992–7 in Calcutta, India. Epidemiol Infect. 2000;124:393–9.
Garg P, Sinha S, Chakraborty R, Bhattacharya SK, Nair GB, Ramamurthy T, et al. Emergence of fluoroquinolone-resistant strains of Vibrio cholerae O1 biotype El Tor among hospitalised patients with cholera in Calcutta, India. Antimicrob Agents Chemother. 2001;45:1605–6.
Das S, Shukla G. Divesity of Vibrio cholera strains isolated in Delhi, India during 1992–2000. J Health Popul Nutr. 2005;23(1):44–51.
Sjolund-Karlsson M, Reimer A, Folster J, Walker M, Dahourou G, Batra D, et al. Drug-Resistance Mechanisms in Vibrio cholerae O1 Outbreak Strain, Haiti, 2010. Emerg Infect Dis. 2011;17(11):2151–4.
Dzotsi EK, Dongdem AZ, Boateng G, Antwi L, Owusu-Okyere G, Nartey DB, et al. Surveillance of Bacterial Pathogens of Diarrhoea in Two Selected Sub Metros Within the Accra Metropolis. Ghana Med J. 2015;49(2):65–71.
Parija SC, Mandal J. Dinoop KP increasing antimicrobial resistance of Vibrio cholera O1 biotype Eltor strains isolated in tertiary care Centre in India. J Health Popul Nutr. 2012;30(1):12–6.
Mala E, Oberoi A, Alexander VS. Vibrio isolates from cases of acute diarrhea and their antimicrobial susceptibility pattern in a tertiary care hospital. Int J Basic Appl Sci. 2014;3(1):35–7.
Klontz EH, Das SK, Ahmed D, Ahmed S, Chisti MJ, Malek MA, et al. Long-Term Comparison of Antibiotic Resistances in Vibrio cholerae O1 and Shigella spp. Between Urban and Rural Bangladesh. Clin Infect Dis. 2014;58(9):e133–6.
Bhattacharya K, Kanungo S, Sur D, Sarkar BL, Manna B, Lopez AL, et al. Tetracycline Resistant Vibrio cholerae O1, Kolkata, India. Emerg Infect Dis. 2011;17(3):568–9.
Shrestha UT, Adhikari N, Maharjan R, Banjara MR, Rijal KR, Basnyat SR, Agrawal VP. Multidrug resistant Vibrio cholerae O1 from clinical and environmental samples in Kathmandu city. BMC Infect Dis. 2015;15:104.
Acknowledgements
We acknowledge all the laboratory staffs of the AMR participating laboratories for their contribution in isolation, identification and sample transport.
Disclosure
The interim findings of the paper were presented as oral presentation at the Joint International Tropical Medicine Meeting, 2017.
Funding
The study was not funded from any external source or funding agency.
Author information
Authors and Affiliations
Contributions
JA: Conception and design of the research, SA: Identification and Laboratory testing, BU: Data Analysis and interpretation, GS: Edited and proofread the document, PK: Critically reviewed the manuscript and final approval of the version, PR: Final editing and approval of the draft, NR: Finalization and drafting the manuscript, All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Data for this study consisted of anonymised laboratory results devoid of individual patient information or identifiers. This study was part of the routine AMR surveillance; hence ethical approval and individual informed consent were not necessary.
Consent for publication
Permission to publish the compiled data was obtained from participating laboratories and National Public Health Laboratory.
Competing interests
The authors declare that they have no competing interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Rijal, N., Acharya, J., Adhikari, S. et al. Changing epidemiology and antimicrobial resistance in Vibrio cholerae: AMR surveillance findings (2006–2016) from Nepal. BMC Infect Dis 19, 801 (2019). https://doi.org/10.1186/s12879-019-4432-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-019-4432-2