Abstract
Introduction
Genome-wide association studies, focusing primarily on unilateral breast cancer, have identified single nucleotide polymorphisms (SNPs) in a number of genomic regions that have alleles associated with a significantly increased risk of breast cancer. In the current study we evaluate the contributions of these previously identified regions to the risk of developing contralateral breast cancer. The most strongly disease-associated SNPs from prior studies were tested for association with contralateral breast cancer. A subset of these SNPs, selected upon their main effects on contralateral breast cancer risk was further evaluated for interaction with treatment modalities and estrogen receptor (ER) status.
Methods
We genotyped 21 SNPs in 708 women with contralateral breast cancer and 1394 women with unilateral breast cancer who serve as the cases and controls in the Women's Environment, Cancer and Radiation Epidemiology (WECARE) Study. Records of treatment and ER status were available for most of WECARE Study participants. Associations of SNP genotypes and risk for contralateral breast cancer were calculated with multivariable adjusted conditional logistic regression methods.
Results
Multiple SNPs in the FGFR2 locus were significantly associated with contralateral breast cancer, including rs1219648 (per allele rate ratio (RR) = 1.25, 95%CI = 1.08-1.45). Statistically significant associations with contralateral breast cancer were also observed at rs7313833, near the PTHLH gene (per allele RR = 1.26, 95%CI = 1.08-1.47), rs13387042 (2q35) (per allele RR = 1.19, 95%CI = 1.02-1.37), rs13281615 (8q24) (per allele RR = 1.21, 95%CI = 1.04-1.40), and rs11235127 near TMEM135 (per allele RR = 1.26, 95%CI = 1.04-1.53). The A allele of rs13387042 (2q35) was significantly associated with contralateral breast cancer in ER negative first tumors while the A allele of rs11235127 (near TMEM135) was significantly associated with contralateral breast cancer in ER positive first tumors. Although some SNP genotypes appeared to modify contralateral breast cancer risk with respect to tamoxifen treatment or particular radiation doses, trend tests for such effects were not significant.
Conclusions
Our results indicate that some common risk variants associated with primary breast cancer also increase risk for contralateral breast cancer, and that these risks vary with the ER status of the first tumor.
Similar content being viewed by others
Introduction
Patients with breast cancer are two to five times more likely to develop a second primary cancer in the contralateral breast than are unaffected women to develop an initial breast cancer [1–4]. Established risk factors for asynchronous second primary contralateral breast cancer (CBC) include those suggesting a genetic basis, such as early age at diagnosis or family history [1], as well as exogenous factors such as the treatment for the first breast cancer [5, 6]. Prior genome-wide association studies (GWASs) have identified regions of the genome containing single-nucleotide polymorphisms (SNPs) with alleles that are associated with an increased risk for breast cancer [7–9]. Many of these SNPs were also reported to be associated with estrogen receptor (ER)-positive breast cancer [9–11]. For the most part, the specific causative variants in these regions and their mechanisms of action remain to be elucidated. In the present study, we evaluated the contributions of these identified loci to risk for CBC.
Our study builds on an existing, population-based, epidemiologic study of CBC: the Women's Environment, Cancer, and Radiation Epidemiology (WECARE) Study. The WECARE study is a nested case-control study in which women with CBC (n = 708) serve as cases and women with unilateral breast cancer (UBC) (n = 1,394) serve as controls [12]. This study population is well characterized for treatment-related exposures, including radiation therapy (RT) (including dose to the contralateral breast), chemotherapy, and hormonal therapy, enabling examination of the interaction of genetic risk factors and cancer therapy. Previous reports from the WECARE Study have shown that RT for a first cancer was associated with an increased risk of CBC [6] but that chemotherapy and tamoxifen treatment were associated with a lower risk of CBC [5]. These treatment effects were further modulated by genetic background. For example, missense variants in the ATM gene were associated with an increased risk of CBC in women who were treated with radiation for their first primary breast tumors [13].
In the present study, we genotyped 21 SNPs representing 18 genomic regions in the WECARE Study population. These SNPs were selected on the basis of their having attained either genome-wide significant evidence or suggestive evidence of association with breast cancer in a prior GWAS that included at least one replication in a separate population. Furthermore, we assessed the interaction of these SNPs with radiation dose to the contralateral breast, chemotherapy, tamoxifen therapy, and ER status.
Materials and methods
Study population
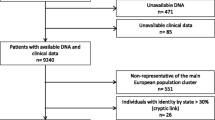
The WECARE Study is a multicenter, population-based, nested case-control study of 708 cases (women with asynchronous bilateral breast cancer) and 1,394 controls (women with UBC) recruited through five population-based cancer registries: one registry covering all of Denmark and, in the US, one registry covering Iowa, two registries covering three counties in Southern California, and one registry covering three counties in Washington State. The controls were individually matched 2:1 to cases on date and age at diagnosis of the first primary breast tumors, race, and registry region and were counter-matched on registry-reported RT. Each case-control triplet consists of two women who received RT and one who did not, maximizing informativeness for radiation exposure [12]. For 12 cases with only one matched control, matched pairs consisted of one exposed and one unexposed woman [12]. The WECARE Study protocol was approved by the institutional review board at each site and the ethics committee system in Denmark, and informed consent was obtained from all participants.
Treatments and phenotypes
Medical records were retrieved to obtain detailed information on the treatment of the first breast cancer (chemotherapy, hormonal therapy, and radiotherapy) during the at-risk period (time between first and second primary diagnoses) and other tumor characteristics, including ER status [5, 6].
Radiation dosimetry
Estimated absorbed radiation doses to various specific contralateral breast locations were reconstructed for each treatment regimen by using tissue-equivalent phantoms and modeling procedures as described previously [6, 12]. Dosage information was available for 604 to 607 cases and 1,184 to 1,195 controls, depending on the number of genotyped individuals per SNP.
Single-nucleotide polymorphism selection
Twenty-one SNPs were selected from among those most associated with breast cancer in three prior GWASs [7–9]. All of these prior studies included at least one replication in a separate population. Nine of the SNPs were associated with breast cancer in prior studies at genome-wide significance levels (P < 10-7). These were rs2981582, rs12443621, rs8051542, rs889312, rs3817198, and rs13281615 from Easton and colleagues [7]; rs1219648 from Hunter and colleagues [8]; rs13387042 from Stacey and colleagues [9]; and rs3803662 reported in both Stacey and colleagues [9] and Easton and colleagues [7]. The remaining 12 SNPs were described as showing evidence of association (global P = 0.001 in three populations) in Easton and colleagues [7] and were listed in the supplemental materials of those authors.
Genotyping
Multiplex SNP genotyping was carried out with the Illumina Golden Gate™ assay on the Sentrix Array Matrix scanned with the Bead Array Reader (Illumina Inc., San Diego, CA, USA). To avoid any bias in the calling of genotypes, all DNA samples were coded and laboratory personnel were blinded to case-control status. A blinded 24% re-sampling was carried out for quality control purposes and these samples were interspersed throughout the plates. Negative controls lacking DNA were included on each plate. Results from a subset of SNPs previously genotyped by an individual SNP method, the MGB Eclipse™ Probe System (Epoch Biosciences, ELITech Group, Paris, France), were concordant with the multiplex genotyping results. Samples with more than 10% missing genotypes and SNPs with more than 5% missing genotypes were excluded from analysis. Although Hardy-Weinberg equilibrium may not strictly apply given that all participants in the study were affected with breast cancer, SNPs deviating from Hardy-Weinberg equilibrium with a P value of less than 0.001 were also excluded.
Haplotyping
Common SNPs (minor allele frequency of greater than 0.1) in the FGFR2 intron 2 region near SNP rs2981582 were selected from the HapMap Project [14]. The Tagger program within Haploview [15] was used to select SNPs informative for the region with an r2 of greater than 0.8. The program PLINK [16] was used to estimate phase. These tagging SNPs were genotyped with the MGB Eclipse™ Probe System. In the haplotype analysis, the referent group for each haplotype consists of those without that haplotype.
Statistical analysis
The WECARE Study is based on a standard nested case-control design with the additional feature of counter-matching on radiotherapy status. All analyses were conducted with multivariable adjusted conditional logistic regression methods, incorporating an 'offset term' or weight to adjust appropriately for the counter-matched sampling [12]. For the SNP main effects analysis, P value for trend was from the log-additive model. The test for trend in Table 1 assessed the hypothesis that CBC risk increases with radiation dose and risk allele 'dose'. Trend tests had 1 degree of freedom. All analyses were performed with SAS (Statistical Analysis System) (SAS Institute Inc., Cary, NC, USA).
Results
The risk of CBC associated with each of the 21 SNPs from 18 genomic regions selected from prior GWASs of UBC [7–9] is shown in Table 2. We observed statistically significant associations with CBC at four of nine SNPs previously reported to be associated with breast cancer at genome-wide significance levels (P < 10-7): rs2981582 (per-allele rate ratio (RR) = 1.20, 95% confidence interval (CI) = 1.04 to 1.40) and rs1219648 (per-allele RR = 1.25, 95% CI = 1.08 to 1.45) at 10q26 in the FGFR2 gene, rs13281615 in the 8q24 region (per-allele RR = 1.21, 95% CI = 1.04 to 1.40), and rs13387042 in the 2q35 region (per-allele RR = 1.19, 95% CI = 1.02 to 1.37). We observed statistically significant associations with CBC at two of the remaining 12 SNPs: rs7313833, located on chromosome 12p in the vicinity of the PTHLH gene (per-allele RR = 1.26, 95% CI = 1.08 to 1.47), and rs11235127 on chromosome 11q14.2 near TMEM135 (per-allele RR = 1.26, 95% CI = 1.04 to 1.53). Excluding BRCA1/2 carriers (who constitute 5.4% of the WECARE Study population [17, 18]) from the present analysis did not markedly alter the results for any of the SNPs tested here (data not shown).
To refine the observed association between CBC and SNPs in the vicinity of FGFR2, we genotyped five haplotype-tagging SNPs, in addition to rs2981582, in all 2,102 available WECARE Study participants and found alleles at four of the six SNPs that were individually associated with risk of CBC (Additional file 1, Table S1). Whereas the risk alleles at these SNPs occurred individually on a number of haplotypes, they occurred together on only two, of which only one was significantly associated with an increased risk of CBC (Additional file 1, Table S2).
For SNPs with a P value of less than 0.1 for main effects in Table 2, we carried out additional exploratory analyses considering the effect on CBC risk of treatment or tumor characteristics of the first cancer. Previous studies have shown that SNP alleles with main effects on the risk for breast cancer can sometimes be associated with specific subtypes of breast cancer [10, 11, 19]. In Table 3, the association of SNPs with CBC was assessed for heterogeneity with respect to the ER status of the first primary tumor. The risk variant at rs13387402 (2q35) was associated with CBC only in individuals with ER-negative first tumors (interaction P value = 0.0008). The risk variant at rs11235127 (near TMEM135) was associated with CBC only in individuals with ER-positive first tumors (interaction P value of not more than 0.01).
There were no significant modifying effects of SNP genotype on CBC risk associated with radiation dose to the contralateral breast for those patients receiving RT for their first cancer (Table 1). Although individual genotypes for SNPs rs3803662 near TOX3 (TNRC9), rs7313833 near PTHLH, and rs2107425 (H19) appeared to be associated with increased risk for CBC in particular dose-allele combinations (for example, for rs7313833 minor allele homozygotes at 0.01 to 0.99 Gy radiation, RR = 2.3, 95% CI = 1.1 to 4.9), there were no obvious trends by either gene or radiation dose. Finally, none of the SNPs studied significantly modified the effects of tamoxifen use or chemotherapy (data not shown) on the incidence of CBC.
Discussion
Breast cancer GWASs have tended to focus on high-risk individuals such as women from multiple-case breast cancer families or with early age at onset. The present study extends findings from these studies by examining another group of genetically predisposed women, those with CBC, and considers important treatment co-factors such as RT, chemotherapy, and hormonal exposures as well as ER status of the first tumor.
We observed nominally significant associations between CBC and SNPs in three regions previously associated with breast cancer, 10q26 (FGFR2), 8q24, and 2q35, all of which had been previously replicated in studies of UBC [20–28]. Rs13387042 (2q35) was also more strongly associated with bilateral than unilateral disease in a recent meta-analysis [28].
Our initial genotyping data revealed an association between an SNP in the FGFR2 gene (rs2981582) and CBC with an RR of a magnitude similar to that observed in the original GWAS report [7]. Further mapping of the FGFR2 intron 2 region with haplotype-tagging SNPs in the WECARE Study population suggested that increased risk for CBC was not restricted to rs2981582 but was associated with a single haplotype, carrying risk alleles at multiple SNPs, consistent with other reports [29, 30].
Among SNPs genotyped in the WECARE Study which were as yet unconfirmed in other studies, the strongest associations with CBC risk were at rs7313833 on chromosome 12p11.22 and at rs11235127 on chromosome 11q14.2. For both of these SNPs, risk was particularly elevated in homozygotes for the minor allele. Although these two SNPs were among the 30 most significantly associated with breast cancer risk in a prior GWAS [7], the associations had not been previously replicated in other study populations [31]. The SNP rs7313833 falls within a small region that is very highly conserved among mammals but that provides no evidence of being a coding region itself. The nearest flanking genes are PTHLH, parathyroid hormone-like hormone, which is located 28 kb centromeric to the reference SNP, and KLHDC5, Kelch domain-containing protein 5, which is located 127 kb telomeric to the reference SNP. In HapMap CEU (Utah residents with Northern and Western European ancestry from the Centre d'Etude du Polymorphism Humain collection) { data, rs7313833 is contained within a region of linkage disequilibrium extending in the centromeric direction to include the relatively small PTHLH gene. PTHLH has a broad range of functions, including roles in mammary gland development and tumor progression [32, 33], making it a potential candidate gene. Annotation of KLHDC5 suggests no obvious connection to breast development or breast cancer.
On chromosome 11, rs11235127 is not located in an evolutionarily conserved region, but there are several uncharacterized, apparently spliced ESTs (expressed sequence tags) in the region. The nearest annotated gene is TMEM135, which is located 54 kb centromeric to the reference SNP and which encodes a transmembrane protein that is not well characterized. The nearest gene in the telomeric direction is RAB38, a Ras family member that is expressed exclusively in melanocytes and that is located more than 700 kb away. We considered TMEM135 to be a candidate gene since studies in the mouse indicate that it is transcriptionally regulated upon treatment with tamoxifen, an estrogen analog and therapeutic agent that is associated with a decreased risk of CBC in the WECARE Study population [34]. However, we observed no statistically significant modifying effect of alleles at rs11235127, the SNP located near TMEM135, or any other SNP studied on the effect of tamoxifen on CBC risk.
In WECARE Study participants who had received RT, the mean radiation dose to the contralateral breast during treatment was 1.1 Gy. In our previous studies of this population, we observed that women who were under 40 years of age and who received more than 1.0 Gy of absorbed dose to a specific quadrant of the contralateral breast had a significant, 2.5-fold greater risk for CBC developing in that quadrant than unexposed women [6]. The presence of specific variants in the ATM gene further increased the risk of CBC specifically in those women who received RT for their first cancer. Accordingly, in the present study, we assessed whether radiation dose-associated CBC risk was modified by genotype; owing to small sample numbers in individual categories, we could not further stratify by age. Overall, the tests for interaction were not statistically significant, although some specific genotype-dose combinations were. For rs3803662 near the TOX3 gene, homozygotes for the reference allele had a statistically significantly increased risk for CBC with radiation dose, whereas carriers of the minor allele did not show statistically significantly increased risk of CBC or dose response. Currently, the function of TOX3 is not well understood and its relevance to radiation response is not clear. The only evident connection between TOX3 and breast cancer is that the gene has been implicated in breast tumor metastasis to bone [35]. Homozygotes for the risk allele at rs7313833, near PTHLH, who received RT had a significantly increased risk of CBC. Interestingly, PTHLH has also been implicated in breast tumor metastasis to bone in a mouse model [32]. Owing to a report that genotype-associated breast cancer risk varied significantly by occupational radiation dose (mean of less than 0.05 Gy) for rs2107425 in H19 [36], we evaluated CBC risk by radiation dose (mean of 1.1 Gy) and rs2107425 genotype. Heterozygotes who received at least 1.0 Gy were at increased risk for CBC, but there was no statistically significant trend with dose and genotype.
Previous studies have assessed whether SNP associations with breast cancer vary with ER status: rs2981582 (in FGFR2), rs3803662 in (TOX3), rs13387042 (in 2q35), and rs13281615 (in 8q24) were more strongly associated with ER-positive disease than ER-negative disease [9, 10, 27]. In the WECARE Study, only rs13387042 (in 2q35) and rs11235127 (near TMEM135) associations with CBC risk displayed statistically significant heterogeneity with regard to ER status. In our study, 27.4% of the CBC cases had an ER-negative first primary tumor and 23.6% of the UBCs were ER-negative overall [17]. The A allele at rs13387042 was associated with increased risk of CBC in participants with ER-negative first tumors. Although an earlier study reported this allele to be associated with ER-positive disease [9], more recent meta-analyses have observed significant associations with both ER-positive and ER-negative disease [27, 37]. Differences in results between the present study and previous studies could reflect specificity to CBC. No previous study has examined rs11235127 (near TMEM135) with regard to ER status.
While the WECARE Study was designed to examine potential interaction effects of genetic risk factors with treatment exposures and tumor characteristics, there are limitations to its application in the context of common risk factors with modest effects. The WECARE Study does not include an unaffected (non-cancer) control population; controls in the study are women with diagnosed UBC. Although women with CBC represent a high-risk breast cancer group, the power to detect main effects of common risk alleles on CBC is limited by the likely enrichment for these same alleles among the UBC patients who serve as controls in the study.
Conclusions
Our study demonstrates that a subset of common variants with moderate effects on breast cancer risk is also associated with an increased risk for CBC. In particular, we identified nominally significant associations with CBC for six of 21 SNPs implicated in prior GWASs of UBC. Two of these SNPs were significantly associated with CBC risk, depending on ER status of the first tumor. Particular combinations of SNP genotypes with radiation dose were associated with increased risk for CBC, although there were no statistically significant interactions with increasing dose and genotype. These findings underscore the need to consider breast cancer tumor characteristics and treatment modalities when examining the risk associated with individual SNPs in order to help characterize their role in breast carcinogenesis.
WECARE Study Collaborative Group
Memorial Sloan-Kettering Cancer Center (New York, NY): Jonine L Bernstein (WECARE Study principal investigator), Colin Begg, Marinela Capanu, Xiaolin Liang, Jennifer Brooks, and Anne R Reiner
City of Hope (Duarte, CA) (some work performed at the University of Southern California, Los Angeles, CA): Leslie Bernstein and Laura Donnelly-Allen
Danish Cancer Society (Copenhagen, Denmark): Jørgen H Olsen, Michael Andersson, Lisbeth Bertelsen, Per Guldberg, and Lene Mellemkjær
Fred Hutchinson Cancer Research Center (Seattle, WA): Kathleen E Malone
International Epidemiology Institute (Rockville, MD) and Vanderbilt University (Nashville, TN): John D Boice Jr.
Lund University (Lund, Sweden): Åke Borg
Mount Sinai School of Medicine (New York, NY): Barry S Rosenstein and David P Atencio
National Cancer Institute (Bethesda, MD): Daniela Seminara
New York University (New York, NY): Roy E Shore
Norwegian Radium Hospital and University of Oslo (Oslo, Norway): Anne-Lise Børresen-Dale
Translational Genomics Research Institute (TGen) (Phoenix, AZ): David Duggan and Ashley Siniard
University of California at Irvine (Irvine, CA): Hoda Anton-Culver (sub-contract principal investigator) and Joan Largent
University of California at Los Angeles (Los Angeles, CA): Richard A Gatti
University of Iowa (Iowa City, IA): Charles F Lynch
University of Southern California (Los Angeles, CA): Robert W. Haile, Graham Casey, Daniel Stram, Duncan C Thomas, Jane Figueiredo, Shanyan Xue, Nianmin Zhou, and Anh T Diep
University of Southern Maine (Portland, ME): W Douglas Thompson
The University of Texas, M.D. Anderson Cancer Center (Houston, TX): Marilyn Stovall and Susan Smith
University of Virginia (Charlottesville, VA) (some work performed at Benaroya Research Institute at Virginia Mason, Seattle, WA): Patrick Concannon, Sharon N Teraoka, Eric R Olson, and Nirasha Ramchurren
Abbreviations
- CBC:
-
contralateral breast cancer
- CI:
-
confidence interval
- ER:
-
estrogen receptor
- GWAS:
-
genome-wide association study
- RR:
-
rate ratio
- RT:
-
radiation therapy
- SNP:
-
single-nucleotide polymorphism
- UBC:
-
unilateral breast cancer
- WECARE:
-
Women's Environment: Cancer: and Radiation Epidemiology.
References
Bernstein J, Thompson W, Risch N, Holford T: Risk factors predicting the incidence of second primary breast cancer among women diagnosed with a first primary breast cancer. Am J Epidemiol. 1992, 136: 925-936.
Li CI, Malone KE, Porter PL, Daling JR: Epidemiologic and molecular risk factors for contralateral breast cancer among young women. Br J Cancer. 2003, 89: 513-518. 10.1038/sj.bjc.6601042.
Berrington de Gonzalez A, Curtis RE, Gilbert E, Berg CD, Smith SA, Stovall M, Ron E: Second solid cancers after radiotherapy for breast cancer in SEER cancer registries. Br J Cancer. 2010, 102: 220-226. 10.1038/sj.bjc.6605435.
Curtis RE, Ron E, Hankey BF, Hoover RN: New malignancies following breast cancer. New malignancies Among Cancer Survivors: SEER Cancer Registries 1973-2000. Edited by: Curtis RE, Freedman DM, Ron E, Ries LAG, Hacker DG, Edwards BK, Tucker MA, Fraumeni JF Jr. 2006, Bethesda MD: US Department of Health and Human Services, National Institutes of Health, National Cancer Institute, 181-205.
Bertelsen L, Bernstein L, Olsen JH, Mellemkjaer L, Haile RW, Lynch CF, Malone KE, Anton-Culver H, Christensen J, Langholz B, Thomas DC, Begg CB, Capanu M, Ejlertsen B, Stovall M, Boice JD, Shore RE, Women's Environment, Cancer and Radiation Epidemiology Study Collaborative Group, Bernstein JL: Effect of systemic adjuvant treatment on risk for contralateral breast cancer in the Women's Environment, Cancer and Radiation Epidemiology Study. J Natl Cancer Inst. 2008, 100: 32-40. 10.1093/jnci/djm267.
Stovall M, Smith SA, Langholz BM, Boice JD, Shore RE, Andersson M, Buchholz TA, Capanu M, Bernstein L, Lynch CF, Malone KE, Anton-Culver H, Haile RW, Rosenstein BS, Reiner AS, Thomas DC, Bernstein JL, Women's Environmental, and Radiation Epidemiology Study Collaborative Group: Dose to the contralateral breast from radiation therapy and risk of second primary breast cancer in the WECARE Study. Int J Radiat Oncol Biol Phys. 2008, 72: 1021-1030. 10.1016/j.ijrobp.2008.02.040.
Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, SEARCH collaborators, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, et al: Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007, 447: 1087-1093. 10.1038/nature05887.
Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Hoover RN, Thomas G, Chanock SJ: A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007, 39: 870-874. 10.1038/ng2075.
Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, Masson G, Jakobsdottir M, Thorlacius S, Helgason A, Aben KK, Strobbe LJ, Albers-Akkers MT, Swinkels DW, Henderson BE, Kolonel LN, Le Marchand L, Millastre E, Andres R, Godino J, Garcia-Prats MD, Polo E, Tres A, Mouy M, Saemundsdottir J, Backman VM, Gudmundsson L, Kristjansson K, Bergthorsson JT, Kostic J, et al: Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007, 39: 865-869. 10.1038/ng2064.
Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, Bojesen SE, Nordestgaard BG, Axelsson CK, Arias JI, Milne RL, Ribas G, González-Neira A, Benítez J, Zamora P, Brauch H, Justenhoven C, Hamann U, Ko YD, Bruening T, Haas S, Dörk T, Schürmann P, Hillemanns P, Bogdanova N, Bremer M, Karstens JH, Fagerholm R, Aaltonen K, Aittomäki K, et al: Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008, 4: e1000054-10.1371/journal.pgen.1000054.
Tapper W, Hammond V, Gerty S, Ennis S, Simmonds P, Collins A, Prospective study of Outcomes in Sporadic versus Hereditary breast cancer (POSH) Steering Group, Eccles D: The influence of genetic variation in 30 selected genes on the clinical characteristics of early onset breast cancer. Breast Cancer Res. 2008, 10: R108-10.1186/bcr2213.
Bernstein JL, Langholz B, Haile RW, Bernstein L, Thomas DC, Stovall M, Malone KE, Lynch CF, Olsen JH, Anton-Culver H, Shore RE, Boice JD, Berkowitz GS, Gatti RA, Teitelbaum SL, Smith SA, Rosenstein BS, Børresen-Dale AL, Concannon P, Thompson WD, WECARE Study: Study design: evaluating gene-environment interactions in the etiology of breast cancer - the WECARE Study. Breast Cancer Res. 2004, 6: R199-R214. 10.1186/bcr771.
Bernstein JL, Haile RW, Stovall M, Boice JD, Shore RE, Langholz B, Thomas DC, Bernstein L, Lynch CF, Olsen JH, Malone KE, Mellemkjaer L, Borresen-Dale AL, Rosenstein BS, Teraoka SN, Diep AT, Smith SA, Capanu M, Reiner AS, Liang X, Gatti RA, Concannon P, WECARE Study Collaborative Group: Radiation exposure, the ATM gene, and contralateral breast cancer in the women's environmental cancer and radiation epidemiology study. J Natl Cancer Inst. 2010, 102: 475-483. 10.1093/jnci/djq055.
International HapMap Project homepage. [http://hapmap.ncbi.nlm.nih.gov/]
Haploview Downloads. [http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/downloads]
PLINK: whole genome association analysis toolset. [http://pngu.mgh.harvard.edu/~purcell/plink/]
Malone KE, Begg CB, Haile RW, Borg A, Concannon P, Tellhed L, Xue S, Teraoka S, Bernstein L, Capanu M, Reiner AS, Riedel ER, Thomas DC, Mellemkjaer L, Lynch CF, Boice JD, Anton-Culver H, Bernstein JL: Population-based study of the risk of second primary contralateral breast cancer associated with carrying a mutation in BRCA1 or BRCA2. J Clin Oncol. 2010, 28: 2404-2410. 10.1200/JCO.2009.24.2495.
Borg A, Haile RW, Malone KE, Capanu M, Diep A, Törngren T, Teraoka S, Begg CB, Thomas DC, Concannon P, Mellemkjaer L, Bernstein L, Tellhed L, Xue S, Olson ER, Liang X, Dolle J, Børresen-Dale AL, Bernstein JL: Characterization of BRCA1 and BRCA2 deleterious mutations and variants of unknown clinical significance in unilateral and bilateral breast cancer: the WECARE sStudy. Hum Mutat. 2010, 31: E1200-1240. 10.1002/humu.21202.
Huijts PE, Vreeswijk MP, Kroeze-Jansema KH, Jacobi CE, Seynaeve C, Krol-Warmerdam EM, Wijers-Koster PM, Blom JC, Pooley KA, Klijn JG, Tollenaar RA, Devilee P, van Asperen CJ: Clinical correlates of low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 in a Dutch cohort of incident breast cancer cases. Breast Cancer Res. 2007, 9: R78-10.1186/bcr1793.
Gold B, Kirchhoff T, Stefanov S, Lautenberger J, Viale A, Garber J, Friedman E, Narod S, Olshen AB, Gregersen P, Kosarin K, Olsh A, Bergeron J, Ellis NA, Klein RJ, Clark AG, Norton L, Dean M, Boyd J, Offit K: Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA. 2008, 105: 4340-4345. 10.1073/pnas.0800441105.
Liang J, Chen P, Hu Z, Zhou X, Chen L, Li M, Wang Y, Tang J, Wang H, Shen H: Genetic variants in fibroblast growth factor receptor 2 (FGFR2) contribute to susceptibility of breast cancer in Chinese women. Carcinogenesis. 2008, 29: 2341-2346. 10.1093/carcin/bgn235.
Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, Jakobsdottir M, Bergthorsson JT, Gudmundsson J, Aben KK, Strobbe LJ, Swinkels DW, van Engelenburg KC, Henderson BE, Kolonel LN, Le Marchand L, Millastre E, Andres R, Saez B, Lambea J, Godino J, Polo E, Tres A, Picelli S, Rantala J, Margolin S, Jonsson T, Sigurdsson H, Jonsdottir T, Hrafnkelsson J, et al: Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008, 40: 703-706. 10.1038/ng.131.
Hemminki K, Müller-Myhsok B, Lichtner P, Engel C, Chen B, Burwinkel B, Försti A, Sutter C, Wappenschmidt B, Hellebrand H, Illig T, Arnold N, Niederacher D, Dworniczak B, Deissler H, Kast K, Gadzicki D, Meitinger T, Wichmann HE, Kiechle M, Bartram CR, Schmutzler RK, Meindl A: Low risk variants FGFR2, TNRC9 and LSP1 in German familial breast cancer patients. Int J Cancer. 2009, 126: 2858-2862.
Fletcher O, Johnson N, Gibson L, Coupland B, Fraser A, Leonard A, dos Santos Silva I, Ashworth A, Houlston R, Peto J: Association of genetic variants at 8q24 with breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2008, 17: 702-705. 10.1158/1055-9965.EPI-07-2564.
Antoniou AC, Spurdle AB, Sinilnikova OM, Healey S, Pooley KA, Schmutzler RK, Versmold B, Engel C, Meindl A, Arnold N, Hofmann W, Sutter C, Niederacher D, Deissler H, Caldes T, Kämpjärvi K, Nevanlinna H, Simard J, Beesley J, Chen X, Kathleen Cuningham Consortium for Research into Familial Breast Cancer, Neuhausen SL, Rebbeck TR, Wagner T, Lynch HT, Isaacs C, Weitzel J, Ganz PA, Daly MB, Tomlinson G, et al: Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet. 2008, 82: 937-948. 10.1016/j.ajhg.2008.02.008.
Mcinerney N, Colleran G, Rowan A, Walther A, Barclay E, Spain S, Jones AM, Tuohy S, Curran C, Miller N, Kerin M, Tomlinson I, Sawyer E: Low penetrance breast cancer predisposition SNPs are site specific. Breast Cancer Res Treat. 2009, 117: 151-159. 10.1007/s10549-008-0235-7.
Reeves GK, Travis RC, Green J, Bull D, Tipper S, Baker K, Beral V, Peto R, Bell J, Zelenika D, Lathrop M, Million Women Study Collaborators: Incidence of breast cancer and its subtypes in relation to individual and multiple low-penetrance genetic susceptibility loci. JAMA. 2010, 304: 426-434. 10.1001/jama.2010.1042.
Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, Hankinson SE, Hutchinson A, Wang Z, Yu K, Chatterjee N, Garcia-Closas M, Gonzalez-Bosquet J, Prokunina-Olsson L, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Diver R, Prentice R, Jackson R, Kooperberg C, Chlebowski R, Lissowska J, et al: A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet. 2009, 41: 579-584. 10.1038/ng.353.
Meyer KB, Maia AT, O'Reilly M, Teschendorff AE, Chin SF, Caldas C, Ponder BA: Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008, 6: e108-10.1371/journal.pbio.0060108.
Udler MS, Meyer KB, Pooley KA, Karlins E, Struewing JP, Zhang J, Doody DR, MacArthur S, Tyrer J, Pharoah PD, Luben R, Bernstein L, Kolonel LN, Henderson BE, Le Marchand L, Ursin G, Press MF, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Kang D, Yoo KY, Noh DY, Ahn SH, Ponder BA, Haiman CA, Malone KE, Dunning AM, Ostrander EA, Easton DF, SEARCH Collaborators: FGFR2 variants and breast cancer risk: fine-scale mapping using African American studies and analysis of chromatin conformation. Hum Mol Genet. 2009, 18: 1692-1703. 10.1093/hmg/ddp078.
Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, Morrison J, Maranian M, Pooley KA, Luben R, Eccles D, Evans DG, Fletcher O, Johnson N, dos Santos Silva I, Peto J, Stratton MR, Rahman N, Jacobs K, Prentice R, Anderson GL, Rajkovic A, Curb JD, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, et al: Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009, 41: 585-590. 10.1038/ng.354.
Guise TA, Yin JJ, Thomas RJ, Dallas M, Cui Y, Gillespie MT: Parathyroid hormone-related protein (PTHrP)-(1-139) isoform is efficiently secreted in vitro and enhances breast cancer metastasis to bone in vivo. Bone. 2002, 30: 670-676. 10.1016/S8756-3282(02)00685-3.
Shen X, Qian L, Falzon M: PTH-related protein enhances MCF-7 breast cancer cell adhesion, migration, and invasion via an intracrine pathway. Exp Cell Res. 2004, 294: 420-433. 10.1016/j.yexcr.2003.11.028.
Fong CJ, Burgoon LD, Williams KJ, Forgacs AL, Zacharewski TR: Comparative temporal and dose-dependent morphological and transcriptional uterine effects elicited by tamoxifen and ethynylestradiol in immature, ovariectomized mice. BMC Genomics. 2007, 8: 151-10.1186/1471-2164-8-151.
Smid M, Wang Y, Klijn JG, Sieuwerts AM, Zhang Y, Atkins D, Martens JW, Foekens JA: Genes associated with breast cancer metastatic to bone. J Clin Oncol. 2006, 24: 2261-2267. 10.1200/JCO.2005.03.8802.
Bhatti P, Doody MM, Alexander BH, Yuenger J, Simon SL, Weinstock RM, Rosenstein M, Stovall M, Abend M, Preston DL, Pharoah P, Struewing JP, Sigurdson AJ: Breast cancer risk polymorphisms and interaction with ionizing radiation among U.S. radiologic technologists. Cancer Epidemiol Biomarkers Prev. 2008, 17: 2007-2011. 10.1158/1055-9965.EPI-08-0300.
Milne RL, Benítez J, Nevanlinna H, Heikkinen T, Aittomäki K, Blomqvist C, Arias JI, Zamora MP, Burwinkel B, Bartram CR, Meindl A, Schmutzler RK, Cox A, Brock I, Elliott G, Reed MW, Southey MC, Smith L, Spurdle AB, Hopper JL, Couch FJ, Olson JE, Wang X, Fredericksen Z, Schürmann P, Bremer M, Hillemanns P, Dörk T, Devilee P, van Asperen CJ, et al: Risk of estrogen receptor-positive and -negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. J Natl Cancer Inst. 2009, 101: 1012-1018. 10.1093/jnci/djp167.
Acknowledgements
This work was supported by National Institutes of Health grants R01 CA129639, R01 CA097397, R01 CA114236, U01 CA083178 (to JB), R01 CA57569 (to PC), and R01 CA112450 (to PC).
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Competing interests
LB is an editor of Breast Cancer Research. All other authors declare that they have no competing interests.
Authors' contributions
SNT participated in study design, data acquisition (molecular genetics) and interpretation, and manuscript drafting and revision. JLB participated in study design, manuscript revision, and study supervision. ASR and MC participated in statistical analysis and interpretation of the data and in manuscript revision. RWH, LB, CFL, MS, SAS, and LM participated in data acquisition and manuscript revision. KEM participated in data acquisition and manuscript revision and provided critical manuscript revision. XL, JM, and XH participated in data analysis and manuscript revision. AS and DD participated in data acquisition (molecular genetics) and manuscript revision. JDB participated in data interpretation and critical manuscript revision. DCT participated in statistical analysis and interpretation and in critical manuscript revision. PC participated in study design, data interpretation, critical manuscript revision, and study supervision. The WECARE Study Collaborative Group participated in recruitment and data acquisition. All authors read and approved the final manuscript.
Electronic supplementary material
13058_2011_2867_MOESM1_ESM.DOC
Additional file 1: Table S1. Risk of Contralateral breast cancer associated with FGFR2 SNPs. Table S2: Risk of Contralateral breast cancer associated with FGFR2 haplotypes. (DOC 40 KB)
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Teraoka, S.N., Bernstein, J.L., Reiner, A.S. et al. Single nucleotide polymorphisms associated with risk for contralateral breast cancer in the Women's Environment, Cancer, and Radiation Epidemiology (WECARE) Study. Breast Cancer Res 13, R114 (2011). https://doi.org/10.1186/bcr3057
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/bcr3057