Abstract
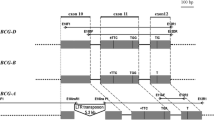
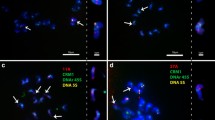
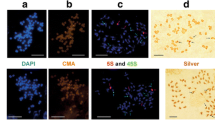
We have cloned and sequenced 115 repeat units of the 5S rDNA genes and spacers from wheat (Triticum) and the polyploid wheat progenitor, Aegilops, and analyzed them together with sequences available in GenBank® (National Center for Biotechnology Information, NCBI, NLM, NIH, Bethesda, Maryland, USA). We were able to assort the sequences into nine orthologous groups which we call unit classes. The following unit classes were assigned to haplomes, and labeled accordingly: long A1, short A1, short A2, long G1, short G1, long D1, short D1, long S1 and short S1. The AA-genome, DD-genome and SS-genome species were each found to contain a long and a short class. The AAGG-genome species T. timopheevii and the AAAtAtGG-genome species T. zhukovskyi, both contain the long A1, long G1 and short G1 unit classes. The AABB-genome species T. turgidum consists of the short A2, a unit class not yet found in T. monococcum, and the long S1 unit class found in the species of Aegilops section Sitopsis. The bread wheat AABBDD-genome contains the long A1, short A2, long D1, long S1 and short G1 unit classes. The presence of the long S1, also demonstrated to occur in both T. turgidum and T. aestivum, supports the hypothesis that the progenitor of the B-haplome in wheat originated in Aegilops section Sitopsis. The presence of the short G1 unit class, i.e. the G-haplome in bread wheat, is unexpected. These new findings are discussed in the light of published findings, especially those relating to 5S DNA loci and evolutionary hypotheses.
Similar content being viewed by others
References
Altschul S.F., Gish W., Miller W., Myers E.W. and Lipman D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410.
Altschul S.F., Stephen F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W. and Lipman D.J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database programs. Nucl. Acids Res. 25: 3389–3402.
Appels R. and Baum B.R. 1991. Evolution of the NOR and 5S DNA loci in the Triticeae. In: Doyle J.J. and Soltis P.S. (Eds.), PlantMolecular Systematics, pp. 92–116. Chapman & Hall, New York.
Appels R., Baum B.R. and Clarke B.C. 1992. The 5S DNA units of bread wheat (Triticum aestivum). Pl. Syst. Evol. 183: 183–194.
Baum B.R. and Appels R. 1992. Evolutionary change at the 5S DNA loci of species in the Triticeae. Plant Syst. Evol. 183: 195–208.
Baum B.R. and Bailey L.G. 1997. The molecular diversity of the 5S rRNA gene in Kengyilia alatavica (Drobov) J.L. Yang, Yen and Baum (Poaceae: Triticeae): potential genomic assignment of different rDNA units. Genome 40: 215–228.
Baum B.R. and Johnson D.A. 1994. The molecular diversity of the 5S rRNA gene in barley (Hordeum vulgare). Genome 37: 992–998.
Baum B.R. and Johnson D.A. 1996. The 5S rRNA gene units in ancestral two-rowed barley (Hordeum spontaneum C. Koch) and bulbous barley (H. bulbosum L.): sequence analysis and phylogenetic relationships with the 5S rDNA units of cultivated barley (H. vulgare L.). Genome 39: 140–149.
Baum B.R. and Johnson D.A. 1998. The 5S rRNA gene in sea barley (Hordeum marinum Hudson sensu lato): sequence variation among repeat units and relationship to the X haplome in barley (Hordeum). Genome 41: 652–661.
Baum B.R. and Johnson D.A. 1999. The 5S rRNA gene in wall barley (Hordeum murinum L. sensu lato): sequence variation among repeat units and relationship to the Y haplome in the genus Hordeum (Poaceae: Triticeae). Genome 42: 854–866.
Baum B.R., Johnson D.A. and Bailey L.G. 1998. Analysis of 5S rDNA units in the Triticeae: the potential to assign sequence units to haplomes. In: Jaradat, A.A. (Ed.), Triticeae III, pp. 85–96. Science Publishers Inc., Enfield, New Hampshire.
Bowden W.M. 1959. The taxonomy and nomenclature of the wheats, barleys, and ryes and their wild relatives. Can. J. Bot. 37: 657–684.
Coulthart M.B., Spencer D.F. and Gray M.W. 1993. Comparative analysis of a recombining-repeat sequence family in the mitochondrial genomes of wheat (Triticum aestivum L.) and rye (Secale cereale L.). Curr. Genet. 23: 255–264.
Cox A.V., Bennett M.D. and Dyer T.A. 1992. Use of the polymerase chain reaction to detect spacer size heterogeneity in plant 5S rRNA gene clusters and to locate such clusters in wheat (Triticum aestivum L.). Theor. Appl. Genet. 83: 684–690.
Dubcovsky J. and Dvořák J. 1995. Genome identification of the Triticum crassum complex (Poaceae) with the restriction patterns of repeated nucleotide sequences. Am. J. Bot. 82: 131–140.
Dvořák J., Zhang H.B., Kota R.S. and Lassner M. 1989. Organization and evolution of the 5S ribosomal RNA gene family in wheat and related species. Genome 32: 1003–1016.
Dvořák J. and Zhang H.B. 1990. Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proc. Natl. Acad Sci. USA 87: 9640–9644.
Dvořák J., di Terlizzi P., Zhang H.B. and Resta P. 1993. The evolution of polyploid wheats: identification of the A genome donor species. Genome 36: 21–31.
Feldman M. 1978. New evidence on the origin of the B genome of wheat. In: Ramanujam, S. (Ed.), Proceedings of the 5th International Wheat Genetics Symposium of the Indian Society, pp 120–132. Genet. Pl. Breeding, New Delhi.
Gerlach W.L. and Dyer T.A. 1980. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucl. Acids Res. 8: 4851–4865.
Gill B.S. and Friebe B. in press. Cytogenetics, phylogeny, and the evolution of cultivated wheats. In: Curtis, B.C. (Ed.), Wheat Production and Improvement. FAO Plant Production and Protection Series.
Hiratsuka J., Shimada H., Whittier H., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C.R., Meng B.Y., Li Y.Q., Kanno K., Nishizawa Y., Hirai A., Shinozaki K. and Sugiura M. 1989. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of cereals. Mol. Gen. Genet. 217: 185–194.
Kimber G. and Feldman M. 1987.Wild wheat: an introduction. Special Report 353. College of Agriculture, University of Missouri, Columbia, Missouri.
Kimber G. and Sears E.R. 1987. Evolution in the genus Triticum and the origin of cultivated wheat. In: Quisenberry, K.S. (Ed.), Wheat and Wheat Improvement. 2nd edition, pp. 154–16.4 Agronomy Monograph No 13, American Society of Agronomy, Madison, WI.
Kimber G. and Tsunewaki K. 1988. Genome symbols and plasma types in the wheat group. In: Miller, T.E. and R.M.D. Koebner (Eds.) Proceedings of the 7th International Wheat Genetics Symposium Vol. 2: pp. 1209–1210. Institute of Plant Science Research.
Leitch I.J. and Heslop-Harrison J.S. 1993. Physical mapping of four sites of 5S rDNA sequences and one site of the "-amylase-2 gene in barley (Hordeum vulgare). Genome 36: 517–523.
Löve Á. 1984. Conspectus of the Triticeae. Feddes Repertorium. 95: 425–521.
Morris R. and Sears E.R. 1967. The cytogenetics of wheat and its relatives. In: Quisenberry, K.S. (Ed.), Wheat and Wheat Improvement. 2nd edn, pp. 19–87. Agronomy Monograph No 13, American Society of Agronomy, Madison, WI.
Mukai Y., Endo T.R. and Gill B.S. 1990. Physical mapping of the 5S rRNA multigene family in common wheat. J. Heredity 81: 290–295.
Nicholas K.B. and Nicholas J.B. Jr. 1997. GeneDoc©: A tool for editing and annotating multiple sequence alignments. V.2.4.002, distributed by the authors.
Page R.D.M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12: 357–358.
Reddy P.R. and Appels R. 1989. A second locus for the multigene family in Secale L.: sequence divergence in two lineages of the family. Genome 32: 456–467.
Sanger F., Nicklen S. and Coulson A.R. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74: 5463–5467.
Thompson J.D., Higgins D.G. and Gibson T.J. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 22: 4673–4680.
Tsunewaki K. 1996. Plasmon analysis as the counterpart of genome analysis. In: Jauhar P.P. (Ed.), Methods of Genome Analysis in Plants, Chap. 16, pp. 271–299. CRC Press, Boca Raton.
Van Campenhout S., Aert R. and Volckaert G. 1998. Orthologous DNA sequence variation among 5S ribosomal RNA gene spacer sequences on homoeologous chromosomes 1B, 1D, and 1R of wheat and rye. Genome 41: 244–255.
Van Slageren M.W. 1994. Wild wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. and Spach) Eig (Poaceae). Wageningen Agric. Papers 94: 7–512.
Yang Y., Baenziger P.S. and Morris R. 1996. Genomic constitution of bread wheat: current status. In: Jauhar, P.P. (Ed.), Methods of Genome Analysis in Plants. Chap. 20, pp. 359–373. CRC Press, Boca Raton.
Yanisch-Perron C., Vieira J. and Messing J. 1985. Improved M13 phage cloning vectors and host strains; nucleotide sequence of M13mp18 and pUC19 vectors. Gene 33: 103–119.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Baum, B.R., Bailey, L.G. The 5S rRNA gene sequence variation in wheats and some polyploid wheat progenitors (Poaceae: Triticeae). Genetic Resources and Crop Evolution 48, 35–51 (2001). https://doi.org/10.1023/A:1011263107219
Issue Date:
DOI: https://doi.org/10.1023/A:1011263107219