Abstract
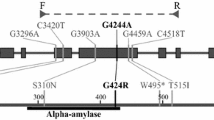
The rice Waxy gene encodes a granule-bound starch synthase (GBSS) necessary for the synthesis of amylose in endosperm tissue. We have previously shown that a CT microsatellite near the transcriptional start site of the GBSS gene can distinguish 7 alleles that accounted for more than 80% of the variation in apparent amylose content in an extended pedigree of 89 US rice cultivars (Oryza sativa L.). Furthermore, all the cultivars with 18% or less amylose were shown to have the sequence AGTTATA at the putative leader intron 5′ splice site, while all cultivars with a higher proportion of amylose had AGGTATA. Here we demonstrate that this single-base mutation reduces the efficiency of GBSS pre-mRNA processing and results in alternate splicing at three cryptic sites. The predominant 5′ splice site in CT18 low-amylose varieties is 93 bp upstream of the splice site used in intermediate and high amylose varieties and is immediately 5′ to the CT microsatellite that we previously demonstrated to be tightly correlated with amylose content. Use of the leader intron 5′ splice site at either -93 or -1 in conjunction with the predominant 3′ splice site results in formation of a small open reading frame 38 bp upstream of the normal ATG and out of frame with it. This open reading frame is not produced when any of the 5′ leader intron splice sites are used in conjunction with an alternate 3′ splice site five bases further downstream which was observed in all rice varieties tested.
Similar content being viewed by others
References
Abler ML, Green PJ: Control of mRNA stability in higher plants. Plant Mol Biol 32: 43–61 (1996).
Ayres NM, McClung AM, Larkin PD, Bligh HFJ, Jones CA, Park WD: Microsatellites and a single nucleotide polymorphism differentiate apparent amylose classes in an extended pedigree of US rice germplasm. Theor Appl Genet 94: 773–781 (1997).
Bligh HFJ, Till R, Jones CA: Amicrosatellite sequence closely linked to the Waxy gene of Oryza sativa. Euphytica 86: 83–85 (1995).
Brown JWS: Arabidopsis intron mutations and pre-mRNA splicing. Plant J 10: 771–780 (1996).
Carle-Urioste JC, Ko CH, Benito M, Walbot V: In vivo analysis of intron processing using splicing-dependent reporter gene assays. Plant Mol Biol 26: 1785–1795 (1994).
Carle-Urioste JC, Brendel V, Walbot V: A combinatorial role for exon, intron, and splice site sequences in splicing in maize. Plant J 11: 1253–1263 (1997).
Chanfreau G., Legrain P, Dujon B, Jacquier A: Interaction between the first and last nucleotides of pre-mRNA introns is a determinant of 3′ splice site selection in S. cerevisiae. Nucl Acids Res 22: 1981–1987 (1994).
Delwiche S, Bean M, Miller R, Webb BD, Williams P: Apparent amylose content of milled rice by near-infrared reflectance spectrophotometry. Cereal Chem 72: 182–187 (1995).
Fu H, Kim SY, Park WD: High level tuber expression and sucrose inducibility of a potato Sus4 sucrose synthase gene require 5′ and 3′ flanking sequences and the leader intron. Plant Cell 7: 1387–1394 (1995).
Futterer J, Hohn T: Translation in plants: rules and exceptions. Plant Mol Biol 32: 159–189 (1996).
Goodall GJ, Filipowicz W: The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58: 473–483 (1989).
Goodall GJ, Filipowicz W: Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J 10: 2635–2644 (1991).
Hinnebusch AG: Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev 52: 248–273 (1988).
Izawa T, Simoto K: Becoming a model plant: the importance of rice to plant science. Trends Plant Sci 1: 95–99 (1996).
Jacobson A, Peltz SW: Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem 65: 693–739 (1996).
Juliano BO: A simplified assay for milled-rice amylose. Cereal Sci Today 16: 334–340 (1971).
Juliano BO: Rice Chemistry and Technology. American Association of Cereal Chemists, St Paul, MN (1985).
Kleffe J, Hermann K, Vahrson W, Wittig B, Brendel V: Logitlinear models for the prediction of splice sites in plant pre-mRNA sequences. Nucl Acids Res 24: 4709–4718 (1996).
Kumar I, Khush GS: Genetic analysis of different amylose levels in rice. Crop Sci 27: 1167–1172 (1987).
Kumar I, Khush GS: Genetic analysis of waxy locus in rice (Oryza sativa L.). Theor Appl Genet 73: 481–488 (1987).
Leuhrsen KL, Walbot V: Intron enhancement of gene expression and the splicing efficiency of introns in maize cells. Mol Gen Genet 225: 81–93 (1991).
Li Y, Ma H, Zhang J, Wang Z, Hong M: Effects of the first intron of rice Waxy gene on the expression of foreign genes in rice and tobacco protoplasts. Plant Sci 108: 181–190 (1995).
Lohmer S, Maddaloni M, Motto M, Salamini F, Thompson RD: Translation of the mRNA of the maize transcriptional activator Opaque 2 is inhibited by upstream open reading frames present in the leader sequence. Plant Cell 5: 65–73 (1993).
McCullough AJ, Lou H, Schuler MA: Factors affecting authentic 5′ splice site selection in plant nuclei. Mol Cell Biol 13: 1323–1331 (1993).
McCullough AJ, Schuler MA: AU-rich intronic elements affect pre-mRNA 5′ splice site selection in Drosophila melanogaster. Mol Cell Biol 13: 7689–7697 (1993).
McCullough AJ, Schuler MA: Intronic and exonic sequences modulate 5′ splice site selection in plant nuclei. Nucl Acids Res 25: 1071–1077 (1997).
Okagaki R: Nucleotide sequence of a long cDNA from the rice waxy gene. Plant Mol Biol 19: 513–516 (1992).
Parker R, Siliciano PG: Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature 361: 660–662 (1993).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY (1989).
Sano Y, Katsumata M, Okumo K: Genetic studies of speciation in cultivated rice. 5. inter-and intraspecific differentiation in the Waxy gene expression of rice. Euphytica 35: 1–9 (1986).
Simpson GJ, Filipowicz W: Splicing of precursors to mRNA in higher plants: mechanism, regulation, and sub-nuclear organisation of the spliceosomal machinery. Plant Mol Biol 32: 1–41 (1996).
Smith CWJ, Porro EB, Patton JG, Nadal-Ginard B: Scanning from an independently specified branchpoint defines the 3′ splice site of mammalian introns. Nature 342: 243–247 (1989).
Smith CWJ, Chu TT, Nadal-Ginard B: Scanning and competition between AG's are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol 13: 4939–4952 (1993).
Waigmann E, Barta A: Processing of chimeric introns in dicot plants: evidence for a close cooperation between 5′ and 3′ splice sites. Nucl Acids Res 20: 75–81 (1992).
Wang Z, Zheng F, Shen G, Gao J, Snustad P, Li M, Zhang J, Hong M: The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J 7: 613–622 (1995).
Webb BD, Bollich C, Johnston T, McIlrath W: Components of rice quality: their identification, methodology, and stage of application in United States breeding programs. In: International Rice Research Institute, Proceedings of the workshop on chemical aspects of rice grain quality, pp. 191–208. International Rice Research Institute, Los Baños, Philippines (1979).
Webb BD: Criteria of rice quality in the United States. In: Juliano BO (ed) Rice Chemistry and Technology, pp. 403–442. American Association of Cereal Chemists, St. Paul, MN (1985).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bligh, H.F.J., Larkin, P.D., Roach, P.S. et al. Use of alternate splice sites in granule-bound starch synthase mRNA from low-amylose rice varieties. Plant Mol Biol 38, 407–415 (1998). https://doi.org/10.1023/A:1006021807799
Issue Date:
DOI: https://doi.org/10.1023/A:1006021807799