Abstract
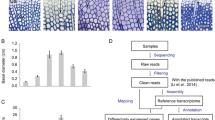
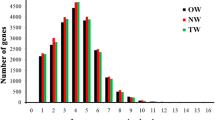
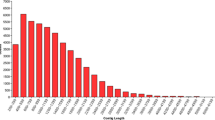
Wood is of critical importance to humans as a primary feedstock for biofuel, fiber, solid wood products, and various natural compounds including pharmaceuticals. The trunk wood of most tree species has two distinctly different regions: sapwood and heartwood. In addition to the major constituents, wood contains extraneous chemicals that can be removed by extraction with various solvents. The composition and the content of the extractives vary depending on such factors as, species, growth conditions, and time of year when the tree is cut. Despite the great commercial and keen scientific interest, little is known about the tree-specific biology of the formation of heartwood and its extractives. In order to gain insight on the molecular regulations of heartwood and its extractive formation, we carried out global examination of gene expression profiles across the trunk wood of black locust (Robinia pseudoacacia L.) trees. Of the 2,915 expressed sequenced tags (ESTs) that were generated and analyzed in the current study, 55.3% showed no match to known sequences. Cluster analysis of the ESTs identified a total of 2278 unigene sets, which were used to construct cDNA microarrays. Microarray hybridization analyses were then performed to survey the changes in gene expression profiles of trunk wood. The gene expression profiles of wood formation differ according to the region of trunk wood sampled, with highly expressed genes defining the metabolic and physiological processes characteristic of each region. For example, the gene encoding sugar transport had the highest expression in the sapwood, while the structural genes for flavonoid biosynthesis were up-regulated in the sapwood-heartwood transition zone. This analysis also established the expression patterns of 341 previously unknown genes.
Similar content being viewed by others
References
Adams, M., Kerlavage, A., Fields, C., and Venter, J. 1993. 3,400 new expressed sequence tags identify diversity of transcripts in human brain. Nat Genet 4: 256–267.
Allona, I., Quinn, M., Shoop, E., Swope, K., Cyr, S., Carlis, J., Riedl, J., Retzel, E., Campbell, M., Sederoff, R., and Whetten, R. 1998. Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA 95: 9693–9698.
Audic, S. and Claverie, J-M. 1997. The significance of digital gene expression profiles. Genome Research 7: 986–995.
Bairoch, A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28: 45–48.
Barker, W., Garavelli, J., Huang, H., McGarvey, P., Orcutt, B., Srinivasarao, G., Xiao, C., Yeh, L.-S.L., Ledley, R., Janda, J., Pfeiffer, F., Mewes, H.-W., Tsugita, A. and Wu, C. 2000. The Protein Information Resource (PIR). Nucleic Acids Res. 28: 41–44.
Baugh, L., Hill, A., Brown, E., and Hunter, C. 2001. Quantitative analysis of mRNA amplification by in vitro transcription. Nuccleic Acid Res. 29: e29.
Benson, D., Karsch-Mizrachi, I., Lipman, D., Ostell, J., Rapp, B., and Wheeler, D. 2000. GenBank. Nucleic Acids Res. 28: 15–18.
Beritognolo, I., Magel, E., Abdel-Latif, A., Charpentier, J., Jay-Allemand, C., and Breton, C. 2002. Expression of genes encoding chalcone synthase, flavanone 3-hydroxylase, and dihydroflavonol 4-reductase correlates with flavanol accumulation during heartwood formation in Juglans nigra L. Tree Physiol. 22: 291–300.
Bevan, M., Bancroft, I., Bent, E., Love, K., Goodman, H., Dean, C., Bergkamp, R., Dirkse, W., Van Staveren, M., Stiekema, W., Drost, L., Ridley, P., Hudson, S.A., Patel, K., Murphy, G., Piffanelli, P., Wedler, H., Wedler, E., Wambutt, R., Weitzenegger, T., Pohl, T.M., Terryn, N., Gielen, J., Villarroel, R., and Chalwatzis, N. 1998. Analysis of 1.9 Mb contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391: 485–488.
Burtin, P., Jay-Allemand, C., Charpentier, J., and Janin, G. 1998 Natural wood colouring process in Juglans sp. (J. nigra, J. regia and hybrid J. nigra 23 × J. regia) depends on native phenolic compounds accumulated in the transition zone between sapwood and heartwood. Trees 12: 258–264.
Carrodus, B., and Triffett, A. 1975. Analysis of respiratory gases in woody stems by mass spectrometry. New Phytol. 74: 243–246.
Cercos, M., Santamaria, S., and Carbonell, J. 1999. Cloning and characterization of TPE4A, a thiol-protease gene induced during ovary senescence and seed germination in pea. Plant Physiol. 119: 1341–1348.
Covitz, P., Smith, L., and Long, S. 1998. Expressed Sequence Tags from a Root-Hair-Enriched Medicago truncatula cDNA library. Plant. Physiol. 117: 1325–1332.
Dazzo, F., and Hubbell, H. 1975. Cross-reactive antigens and lectins as determinants of symbiotic specificity in the Rhizobium–clover association. Appl. Microbiol. 30: 1017–1033.
de Vetten, N., ter Horst, J., van Schaik, H.-P., de Boer, A., Mol, J., and Koes, R. 1999. A cytochrome b5 is required for full activity of flavonoid 3?,5?-hydroxylase, a cytochrome P450 involved in the formation of blue flowers. Proc Natl Acad Sci USA 96: 778–783.
Dent, G., O'Dell, D, and Eberwine, H. 2001. Gene expression profiling in the amygdala: An approach to examine the moleculat substrates of mammalian behavior. Physiol. Behavior 73: 841–847.
Eisen, M., Spellman, P., Brown, P., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868.
Eulgem, T., Rushton, P., Schmelzer, E., Hahlbrock, K., and Somssich, I. 1999 Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 18: 4689–4699.
Ewing, B., Hillier, L., Wendl M., and Green, P. 1998. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185.
Fukuda, H. (1996). Xylogenesis: initiation, prpgression, and cell death. Ann Rev Plant Physiol Plant Mol Biol 47, 299–325.
García-Hernández, M., Murphy, A., and Taiz, L. 1998. Matallothioneins 1 and 2 have distinct but overlapping expreeion patterns in Arabidopsis. Plant Physiol. 118: 387–397.
Hauch, S. and Magel, E. 1998. Extractable activities and protein content of sucrose-phosphate synthase, sucrose synthase and neutral invertase in trunk tissues of Robinia pseudoacacia L. are related to cambial wood production and heartwood formation. Planta 207: 266–274.
Hertzberg, M., Aspeborg, H., Schrader, J., Andersson, A., Erlandsson, R., Blomqvist, K., Bhalerao, R., Uhlén, M., Teeri, T., Lundberg, J., Sundberg, B., Nisson, P., and Sandberg, G. 2001. A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. USA 98: 14732–14737.
Hihara, Y., Kamei, A., Kanehisa, M., Kaplan, A., and Ikeuchi, M. 2001. DNA Microarray Analysis of Cyanobacterial Gene Expression during Acclimation to High Light, Plant Cell 13: 793–806.
Hillinger, C., Holl, W., and Ziegler, H. 1996a. Lipids and lipolytic enzymes in the trunkwood of Robinia pseudoacacia L. during heartwood formation. I. Radial distribution of lipid classes. Trees 10: 366–375.
Hillinger, C., Holl, W., and Ziegler, H. 1996b. Lipids and lipolytic enzymes in the trunkwood of Robinia pseudoacacia L. during heartwood formation. II. Radial distribution of lipases and phospholipases. Trees 10: 376–381.
Hillis W. 1987. Heartwood and the exudates, Spinger, Berlin, Heidelberg, New York.
Ingemarsson, B. 1995. Ethylene effects on peroxidases and cell growth patterns in Picea abies hypocotyl cuttings. Physiol. Plant. 94: 211–218.
Kirst, M., Johnson, A., Retzel, E., van Zyl, L., Craig, D., Li, Z.J., Whetten, R., Baucom, C., Ulrich, E., Hubbard, Kristy, and Sederoff, R. (2002). Quantitative inference in functional genomics of loblolly pine (Pinus taeda L.) using ESTs and microarrays. In Proc. the 10th IAPTC&B Congress. (Ed) I. Vasil. Kluwer Academic Publishers, Dordrecht, Netherlands. Kozlowski, T. and Pallardy, S. (1997). Physiology of woody plants. San Diego, Academic Press.
Lalonde, S., Boles, E., Hellmann, H., Barker, L., Patrick, J., Frommer, W., and Ward, J. 1999. The dual function of sugar carriers: transport and sugar sensing. Plant Cell 11: 707–726.
Lee, M., Kuo, F., Whitmore, G., and Sklar, J. 2000. Importance of replication in microarray gene expression studies: Statistical methods and evidence from repetitive cDNA hybridizations. Proc. Natl. Acad. Sci. USA 97: 9834–9839.
Livesey, F., Furukawa, T., Steffen, M., Church, G., and Cepko, C. 2000 Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Current Biology 10: 301–310.
Lorenz, W. and Dean, J. 2002. SAGE profiling and demonstration of differential gene expression along the axial developmental gradient of lignifying xylem in loblolly pine (Pinus taeda). Tree Physiology 22: 301–310.
Magel, E. 2000. Biochemistry and physiology of heartwood formation. In Molecular and Cell Biology of Wood Formation. Eds. R. Savidge, J. Barnett and R. Napier. BIOS Scientific Publishers, Oxford, pp. 363–376.
Magel, E., Jay-Allemand, C., and Ziegler, H. 1994. Formation of heartwood substances in the stemwood of Robinia pseudoacacia L.: II. Distribution of nonstructural carbohydrates and wood extractives across the trunk. Trees 165–171.
Magel, E. and Hübner, B. 1997 Distribution of phenylalanine ammonia lyase and chalcone synthase within trunks of Robinia pseudoacacia L. Bot. Acta 110: 314–322.
Matamoros, M., Baird, L., Escuredo, P., Dalton, D., Minchin, F., Iturbe-Ormaetxe, I., Rubio, M., Moran, J., Gordon, A., and Becana, M. 1999. Stress-induced legume root nodule senescence. Physiological, biochemical, and structural alterations. Plant Physiol 121: 97–112.
Mauseth, J. 1998.Botany: an introduction to plant biology. Jones and Bartlett Publishers. Sudbury, Massachusetts.
Miller, J., Arteca, R., and Pell, E. 1999. Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol. 20: 1015–1024.
Nelson, N. 1978. Xylem ethylene, phenol oxidising enzymes and nitrogen and heartwood formation in walnut and cherry. Can. J. Bot. 56: 626–634.
Nilsson, M., Wikman, S., and Eklund, L. 2002. Induction of discolored wood in Scots pine (Pinus sylvestris). Tree Physiology 22: 331–338.
Perez-Amador, M., Lidder, P., Johnson, M., Landgraf, J., Wisman, E., and Green, P. 2001. New molecular phenotypes in the dst mutants of Arabidopsis revealed by DNA microarray analysis. Plant Cell 13: 2703–2717.
Reddy, A., and Poovaiah, B. 1990. Molecular cloning and sequencing of a cDNA for an auxin-repressed mRNA: correlation between fruit growth and repression of the auxin-regulated gene. Plant Mol Biol. 14: 127–136.
Roberts, L. and Miller, A. 1983 Is ethylene involved in xylem of differentiation? Plant Physiol. 6: 1–24.
Saxe, H., Ellsworth, D. S., and Heath, J. 1998. Tree and forest functioning in an enriched CO2 atmosphere. New Phytol. 139: 395–436.
Schaffer, R., Landgraf, J., Accerbi, M., Simon, V., Larson, M., and Wisman, E. 2001. Microarray Analysis of Diurnal and Circadian-Regulated Genes in Arabidopsis. Plant Cell 13: 113–123.
Sederoff, R., Kirst, M., Johnson, A., Retzel, E., Whetten, R., Vasques-Kool, J. & O'Malley, D. 2002. Homology of expressed genes in loblolly pine (Pinus taeda L.) with Arabidopsis thaliana. The 10th IAPTC&B Congress, February 23–28, 2002, Orlando, FL.
Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. 2001. Monitoring the Expression Pattern of 1300 Arabidopsis Genes under Drought and Cold Stresses by Using a Full-Length cDNA Microarray. Plant Cell 13: 61–72.
Stafstrom, J., Ripley, B., Devitt, M., and Drake, B. 1998. Dormancy-associated gene expression in pea axillary buds. Cloning and expression of PsDRM1 and PsDRM2. Planta 205: 547–552.
Sterky, F., Regan, S., Karlsson, J., Hertzberg, M., Rohde, A., Holmberg, A., Amini, B., Bhalerao, R., Larsson, M., Villarroel, R., Van Montagu, M., Sandberg, G., Olsson, O., Teeri, T. T., Boerjan, W., Gustafsson, P., Uhlen, M., Sundberg, B., and Lundeberg, J. 1998. Gene discovery in the wood-forming tissues of poplar: analysis of 5, 692 expressed sequence tags. Proc Natl Acad Sci USA 95: 13330–13335.
Steward, C. 1966. Excretion and heartwood formation in living trees. Science 153: 1068–1074.
Uggla, C., Magel, E., Moritz, T., and Sundberg, B. 2001. Function and dynamics of auxin and carbohydrates during early wood/late wood transition in Scots pine. Plant Physiol. 125: 2029–2039.
Wang, E., Miller, L., Ohnmacht, G., Liu, E., and Marincola, F 2000. High-fidelity mRNA amplification for gene profiling. Nat. Biotech 18: 457–459.
Williams, L., Lemoine, R., and Sauer, N. 2000. Sugar transporters in higher plants – a diversity of roles and complex regulation. Trens in Plant Science 5: 283–290.
Wingler, A., von Schaewen, A., Leegood, R., Lea, P., and Quick, W. 1998. Regulation of leaf senescence by cytokinin, sugars, and light. Plant Physiol. 116: 329–335.
Yu, Q., He, M., Lee, N., and Liu, E. (2002). Identification of MYCmediated death response pathways by microarray analysis. J Biol Chem. 277: 13059–13066.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, J., Park, S., Kamdem, D.P. et al. Novel gene expression profiles define the metabolic and physiological processes characteristic of wood and its extractive formation in a hardwood tree species, Robinia pseudoacacia . Plant Mol Biol 52, 935–956 (2003). https://doi.org/10.1023/A:1025445427284
Issue Date:
DOI: https://doi.org/10.1023/A:1025445427284