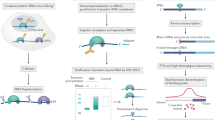
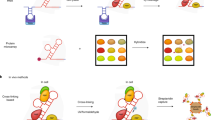
Abstract
RNA-nucleoprotein (RNP) complexes are pivotal to the regulation of gene expression. These multiprotein, multifunctional complexes act as scaffolds, as molecular platforms, as hubs of interaction between the RNA and the proteins. The past decade has seen an exponential increase in the methodologies to identify and functionally characterize these complexes. In this review, we provide an overview of the recent advances in methodologies and the knowledge gained by characterizing these important complexes. This information has provided clues to the role of sequence, and structural determinants for RNA–Protein interaction networks in physiology and pathophysiology. With the recent studies implicating the role of RNA in the formation of biomolecular condensates, their role in health and disease warrants detailed sequence, structure and functional investigations.







Similar content being viewed by others
Abbreviations
- AD:
-
Alzheimers disease
- AD:
-
Autosomal dominant
- ADHD:
-
Attention deficit hyperactive disorder
- ADPD:
-
Attention deficit passive disorder.
- ALS:
-
Amyotrophic lateral sclerosis
- AR:
-
Autosomal recessive
- ALKBH5:
-
AlkB homolog 5 RNA demethylase
- APEX-Seq:
-
APEX-2 mediated cross-linking and sequencing
- APP:
-
Amyloid precursor protein
- ARM:
-
Arginine rich motif
- ASD:
-
Autism spectrum disorder
- ATXN2:
-
Ataxin 2 protein
- BD:
-
Bipolar disorder
- CARIC:
-
Click chemistry assisted RNA-interactome capture
- CCR4/NOT:
-
Carbon-catabolite repression 4-negative on TATA-less
- CHART-MS:
-
Capture hybridization analysis of RNA targets
- ChIRP-MS:
-
Comprehensive identification of RNA-binding proteins by mass spectroscopy
- CLIP:
-
Cross-linking and immunoprecipitation
- CRAC:
-
Cross-linking and cDNA analysis
- CRUIS:
-
CRISPR based RNA based interacting system
- DCP1/DCP2:
-
Decapping protein 1 and 2
- DSRM:
-
Double stranded RNA binding motif
- eCLIP:
-
Enhanced CLIP
- ED:
-
Edmann degradation
- FCS:
-
Fluorescence correlation spectroscopy
- FISH:
-
Fluorescence in-situ hybridization
- FMRP:
-
Fragile X mental retardation protein
- FRAP:
-
Fluorescence recovery after photobleaching
- FRET:
-
Fluorescence resonance energy transfer
- FUS:
-
Fused in sarcoma
- FTD:
-
Frontotemporal dementia
- FTO:
-
Fat mass and obesity-associated
- HD:
-
Huntington’s disease
- HTT:
-
Huntington’s disease gene huntingtin
- hnRNP:
-
Heterogeneous nuclear ribonucleoprotein.
- HTR-SELEX:
-
High-throughput systematic evolution of ligands by exponential enrichment.
- IP:
-
Immunoprecipitation.
- KH Motif:
-
K-Homology motif
- m6A:
-
6-Methyl adenosine
- MATR3:
-
Matrin 3
- MD:
-
Mood disorder
- MDD:
-
Major depressive disorder
- METTL3/14:
-
Methyltransferase like protein 3/14
- MLO:
-
Membraneless organelles
- MS:
-
Mass spectrometry
- Mu:
-
Multifactorial
- NMIA:
-
N-Methylisatoic anhydride
- NMR:
-
Nuclear magnetic resonance spectroscopy
- OMIM:
-
Online Mendalian Inheritancein Man
- PAN2/PAN3:
-
Poly(A) nuclease deadenylation complex
- PAZ:
-
Piwi/Argonaute/Zwille
- PABP:
-
Poly(A) binding protein
- PAR-CLIP:
-
Photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation
- PD:
-
Parkinsons disease
- PIWI:
-
P-element induced wimpy testis
- QKI:
-
Quackinguiable protein 1
- RaPID:
-
RNA–protein interaction detection
- RAP-MS:
-
RNA antisense purification coupled with mass spectrometry
- RNP:
-
Ribonucleoprotein
- RNA:
-
Ribonucleic acid
- RBP:
-
RNA binding protein
- RRM:
-
RNA recognition motif
- RGG:
-
Arginine glycine glycine motif
- RNP-CSI:
-
RNP consensus sequence
- RBD:
-
RNA binding domain
- RBM-45:
-
RNA binding motif 45
- SCA2:
-
Spinocerebellar ataxia 2
- SCZ:
-
Schizophrenia
- SG:
-
Stress granules
- SHAPE:
-
Selective 2′-hydroxyl acylation and primer extension
- SF:
-
Splicing factor
- SMN:
-
Spinal muscular atrophy
- snRNP:
-
Small nuclear ribonucleoprotein
- TARDBP/TDP-43:
-
Transactive response DNA binding protein
- TDP-43:
-
TAR DNA binding protein
- TRAP:
-
Translating ribosome affinity purification
- TRAPP:
-
Total RNA associated protein purification
- TRIBE:
-
Targets of RNA-binding proteins identified by editing
- UPS:
-
Ubiquitin proteosome system
- XLD:
-
X linked dominan
- XNR1:
-
Xenopus nodal related-1
References
Abdelmohsen, K., Srikantan, S., Kuwano, Y., Gorospe, M.: miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. USA 105(51), 20297–20302 (2008)
Agrawal, S., Kuo, P.H., Chu, L.Y., Golzarroshan, B., Jain, M., Yuan, H.S.: RNA recognition motifs of disease-linked RNA-binding proteins contribute to amyloid formation. Sci. Rep. 9, 6171 (2019)
Allfrey, V., Daly, M.M., Mirsky, A.E.: Synthesis of protein in the pancreas. II. The role of ribonucleoprotein in protein synthesis. J. Gen. Physiol. 37, 157–175 (1953)
ATtRACT database: https://attract.cnic.es/index
Aumiller, W.M., Jr., Pir Cakmak, F., Davis, B.W., Keating, C.D.: RNA-based coacervates as a model for membraneless organelles: formation, properties, and interfacial liposome assembly. Langmuir 32, 10042–10053 (2016)
Auweter, S.D., Oberstrass, F.C., Allain, F.H.T.: Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 34, 4943–4959 (2006)
Auweter, S.D., Oberstrass, F.C., Allain, F.H.T.: Solving the structure of PTB in complex with pyrimidine tracts: an NMR study of protein-RNA complexes of weak affinities. J. Mol. Biol. 367, 174–186 (2007)
Bandziulis, R.J., Swanson, M.S., Dreyfuss, G.: RNA-binding proteins as developmental regulators. Genes Dev. 3, 431–437 (1989)
Beach, D.L., Keene, J.D.: Ribotrap : targeted purification of RNA-specific RNPs from cell lysates through immunoaffinity precipitation to identify regulatory proteins and RNAs. Methods Mol. Biol. 419, 69–91 (2008)
Beckmann, B.M., Horos, R., Fischer, B., Castello, A., Eichelbaum, K., Alleaume, A.M., Schwarzl, T., Curk, T., Foehr, S., Huber, W., Krijgsveld, J., Hentze, M.W.: The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 6, 10127 (2015)
Bharathavikru, R., Dudnakova, T., Aitken, S., Slight, J., Artibani, M., Hohenstein, P., Tollervey, D., Hastie, N.: Transcription factor Wilms’ tumor 1 regulates developmental RNAs through 3’ UTR interaction. Genes Dev. 31, 347–352 (2017)
Bharathavikru, R., Dudnakova, T.: Methods to identify and validate WT1-RNA interaction. Methods Mol. Biol. 1467, 197–209 (2016). https://doi.org/10.1007/978-1-4939-4023-3_17
Bhardwaj, A., Myers, M.P., Buratti, E., Baralle, F.E.: Characterizing TDP-43 interaction with its RNA targets. Nucleic Acids Res. 41, 5062–5074 (2013)
Boehr, D.D.: Promiscuity in protein-RNA interactions: conformational ensembles facilitate molecular recognition in the spliceosome: conformational diversity in U2AF 65 facilitates binding to diverse RNA sequences. BioEssays 34(3), 174–180 (2012)
Bokar, J.A., Rath-Shambaugh, M.E., Ludwiczak, R., Narayan, P., Rottman, F.: Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 269(26), 17697–17704 (1994)
Brenner, S., Jacob, F., Meselson, M.: An unsTable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190, 576–581 (1961)
Buckley, P.T., Khaladkar, M., Kim, J., Eberwine, J.: Cytoplasmic intron retention, function, splicing, and the sentinel RNA hypothesis. Wiley Interdiscip. Rev. RNA 5, 223–230 (2014)
Burd, C.G., Matunis, E.L., Dreyfuss, G.: The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol. Cell Biol. 11, 3419–3424 (1991). https://doi.org/10.1128/mcb.11.7.3419-3424
Campbell, Z.T., Bhimsaria, D., Valley, C.T., Rodriguez-Martinez, J.A., Menichelli, E., Williamson, J.R., Ansari, A.Z., Wickens, M.: Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell Rep. 1, 570–581 (2012)
Castello, A., Fischer, B., Frese, C.K., Horos, R., Alleaume, A.M., Foehr, S., Curk, T., Krijgsveld, J., Hentze, M.W.: Comprehensive identification of RNA-binding domains in human cells. Mol. Cell 63, 696–710 (2016)
Chao, F.C.: Dissociation of macromolecular ribonucleoprotein of yeast. Arch. Biochem. Biophys. 70, 426–431 (1957)
Choi, J.M., Holehouse, A.S., Pappu, R.V.: Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 49, 107–133 (2020)
Chu, C., Chang, H.Y.: ChIRP-MS: RNA-directed proteomic discovery. Methods Mol. Biol. 1861, 37–45 (2018)
Conlon, E.G., Manley, J.L.: RNA-binding proteins in neurodegeneration: mechanisms in aggregate. Genes Dev. 31, 1509–1528 (2017)
Cookson, M.R.: RNA-binding proteins implicated in neurodegenerative diseases. Wiley Interdiscip. Rev. RNA (2017). https://doi.org/10.1002/wrna.1397
Corley, M., Burns, M.C., Yeo, G.W.: How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell 78, 9–29 (2020)
Crick, F.H.: On protein synthesis. In: Sanders, F.K. (ed.) Symposia of the Society for Experimental Biology, Number XII: The Biological Replication of Macromolecules, pp. 138–163. Cambridge University Press (1958)
Database of AminoAcid Indices. https://www.genome.jp/aaindex/
Davidson, J.N.: Cytoplasmic ribonucleoproteins. Biochem. J. 39(5), lix–lxi (1945)
Desrosiers, R., Friderici, K., Rottman, F.: Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 71(10), 3971–3975 (1974)
Dexheimer, P.J., Wang, J., Cochella, L.: Two MicroRNAs are sufficient for embryonic patterning in C. elegans. Curr. Biol. 30(24), 5058-5065.e5 (2020)
Dominguez, D., Freese, P., Alexis, M.S., Su, A., Hochman, M., Palden, T., Bazile, C., Lambert, N.J., Van Nostrand, E.L., Pratt, G.A., Yeo, G.W., Graveley, B.R., Burge, C.B.: Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell. 70, 854-867.e9 (2018)
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., Cesarkas, K., Jacob-Hirsch, J., Amariglio, N., Kupiec, M., Sorek, R., Rechavi, G.: Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485(7397), 201–206 (2012)
Elbaum-Garfinkle, S., Kim, Y., Szczepaniak, K., Chen, C.C., Eckmann, C.R., Myong, S., Brangwynne, C.P.: The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA 112(23), 7189–7194 (2015)
Fazal, F.M., Han, S., Parker, K.R., Kaewsapsak, P., Xu, J., Boettiger, A.N., Chang, H.Y., Ting, A.Y.: Atlas of subcellular RNA localization revealed by APEX-Seq. Cell 178, 473-490.e26 (2019)
Fernández, E., Rajan, N., Bagni, C.: The FMRP regulon: from targets to disease convergence. Front. Neurosci. 24(7), 191 (2013)
Ferus, M., Pietrucci, F., Saitta, A.M., Knížek, A., Kubelík, P., Ivanek, O., Shestivska, V., Civiš, S.: Formation of nucleobases in a Miller-Urey reducing atmosphere. Proc. Natl. Acad. Sci. USA 114, 4306–4311 (2017)
Fu, Y., Zhuang, X.: m6A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 16(9), 955–963 (2020)
Gagliardi, M., Matarazzo, M.R.: RIP: RNA immunoprecipitation. Methods Mol. Biol. 1480, 73–86 (2016)
Georgiev, G.P., Samarina, O.P., Mantieva, V.I., Zbarsky, I.B.: Further studies on the nuclear ribonucleoproteins. Biochim. Biophys. Acta 46, 399–400 (1961)
Glazer, R.I., Vo, D.T., Penalva, L.O.: Musashi1: an RBP with versatile functions in normal and cancer stem cells. Front. Biosci. 17(1), 54–64 (2012)
Glisovic, T., Bachorik, J.L., Yong, J., Dreyfuss, G.: RNA-binding proteins and posttranscriptional gene regulation. FEBS Lett. 582, 1977–1986 (2008)
Guillén-Boixet, J., et al.: RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346-361.e17 (2020)
Gupta, A., Gribskov, M.: The role of RNA sequence and structure in RNA–protein interactions. J. Mol. Biol. 409, 574–587 (2011)
Heat mapper. http://heatmapper.ca/
Helwak, A., Kudla, G., Dudnakova, T., Tollervey, D.: Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665 (2013)
Hentze, M., Castello, A., Schwarzl, T., et al.: A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341 (2018)
Holley, R.W., Apgar, J., Everett, G.A., Madison, J.T., Marquisee, M., Merrill, S.H., Penswick, J.R., Zamir, A.: Structure of a ribonucleic acid. Science 147, 1462–1465 (1965)
Huang, R., Han, M., Meng, L., Chen, X.: Transcriptome-wide discovery of coding and noncoding RNAbinding proteins. Proc. Natl. Acad. Sci. USA 115, E3879–E3887 (2018)
Huranova, M., Jablonski, J.A., Benda, A., Hof, M., Stanek, D., Caputi, M.: in vivo detection of RNA-binding protein interactions with cognate RNA sequences by fluorescence resonance energy transfer. RNA 2009(15), 2063–2071 (2009)
Jain, S.S., Tullius, T.D.: Footprinting protein-DNA complexes using the hydroxyl radical. Nat. Protoc. 3, 1092–1100 (2008)
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., Yi, C., Lindahl, T., Pan, T., Yang, Y.G., He, C.: N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7(12), 885–887 (2011)
Jolma, A., Kivioja, T., Toivonen, J., Cheng, L., Wei, G., Enge, M., Taipale, M., Vaquerizas, J.M., Yan, J., Sillanpää, M.J., Bonke, M., Palin, K., Talukder, S., Hughes, T.R., Luscombe, N.M., Ukkonen, E., Taipale, J.: Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 20, 861–873 (2010)
Jolma, A., Zhang, J., Mondragón, E., Morgunova, E., Kivioja, T., Laverty, K.U., Yin, Y., Zhu, F., Bourenkov, G., Morris, Q., Hughes, T.R., Maher, L.J., Taipale, J.: Binding Specificities of human RNA binding proteins towards structured and linear RNA sequences. Genome Res. 30, 962–973 (2020)
Kanaan, N.M., Hamel, C., Grabinski, T., Combs, B.: Liquid-liquid phase separation induces pathogenic tau conformations in vitro. Nat. Commun. 11(1), 2809 (2020)
Kawahara, Y., Mieda-Sato, A.: TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. USA 109, 3347–3352 (2012)
Kim, T.H., Payliss, B.J., Nosella, M.L., Lee, I.T.W., Toyama, Y., Forman-Kay, J.D., Kay, L.E.: Interaction hot spots for phase separation revealed by NMR studies of a CAPRIN1 condensed phase. Proc. Natl. Acad. Sci. USA 118(23), e2104897118 (2021)
Kishore, S., Luber, S., Zavolan, M.: Deciphering the role of RNA-binding proteins in the posttranscriptional control of gene expression. Brief Funct. Genomics 9, 391–404 (2010)
Krause, H., Simmonds, A.: Trap-tagging: a novel method for the identification and purification of RNA–protein complexes. In: Google Patents (2006)
Lafontaine, D.L.J., Riback, J.A., Bascetin, R., Brangwynne, C.P.: The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165–182 (2021)
Lambert, N., Robertson, A., Jangi, M., McGeary, S., Sharp, P.A., Burge, C.B.: RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell 54, 887–900 (2014)
Lancet, D., Zidovetzki, R., Markovitch, O.: Systems protobiology: origin of life in lipid catalytic networks. J. r. Soc. Interface 15, 20180159 (2018)
Le Vay, K., Weise, L.I., Libicher, K., Mascarenhas, J., Mutschler, H.: Templated self-replication in biomimetic systems. Adv. Biosyst. 3, e1800313 (2019)
Lee, H.Y., Haurwitz, R.E., Apffel, A., Zhou, K., Smart, B., Wenger, C.D., Laderman, S., Bruhn, L., Doudna, J.A.: RNA-protein analysis using a conditional CRISPR nuclease. Proc. Natl. Acad. Sci. USA 110, 5416–5421 (2013)
Leppek, K., Stoecklin, G.: An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res. 42, e13 (2014)
Licatalosi, D.D., Mele, A., Fak, J.J., Ule, J., Kayikci, M., Chi, S.W., Clark, T.A., Schweitzer, A.C., Blume, J.E., Wang, X., Darnell, J.C., Darnell, R.B.: HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 (2008)
Licatalosi, D.D., Ye, X., Jankowsky, E.: Approaches for measuring the dynamics of RNA-protein interactions. Wiley Interdiscip. Rev. RNA 11(1), e1565 (2020)
Lin, X., Fonseca, M.A.S., Breunig, J.J., Corona, R.I., Lawrenson, K.: In vivo discovery of RNA proximal proteins via proximity-dependent biotinylation. RNA Biol. 18(12), 2203–2217 (2021)
Littlefield, J.W., Keller, E.B., Gross, J., Zamenick, P.C.: Studies on cytoplasmic ribonucleoprotein particles from the liver of the rat. J. Biol. Chem. 217, 111–123 (1955)
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., Jia, G., Yu, M., Lu, Z., Deng, X., Dai, Q., Chen, W., He, C.: A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10(2), 93–95 (2014)
Lunde, B.M., Moore, C., Varani, G.: RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490 (2007)
Maharana, S., Wang, J., Papadopoulos, D.K., Richter, D., Pozniakovsky, A., Poser, I., Bickle, M., Rizk, S., Guillén-Boixet, J., Franzmann, T.M., Jahnel, M., Marrone, L., Chang, Y.T., Sterneckert, J., Tomancak, P., Hyman, A.A., Alberti, S.: RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360(6391), 918–921 (2018)
Maziuk, B., Ballance, H.I., Wolozin, B.: Dysregulation of RNA binding protein aggregation in neurodegenerative disorders. Front. Mol. Neurosci. 10, 89 (2017)
McMahon, A.C., et al.: TRIBE: hijacking an RNA-editing enzyme to identify cell-specific targets of RNA-binding proteins. Cell 165, 742–753 (2016)
Merino, E.J., Wilkinson, K.A., Coughlan, J.L., Weeks, K.M.: RNA structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231 (2005)
Meyer, K.D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C.E., Jaffrey, S.R.: Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149(7), 1635–1646 (2012)
Michlewski, G., Cáceres, J.F.: RNase-assisted RNA chromatography. RNA 16, 1673–1678 (2010)
Mitrea, D.M., Kriwacki, R.W.: Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 14, 1 (2016)
Mucsi, Z., Hudecz, F., Hollósi, M., Tompa, P., Friedrich, P.: Binding-induced folding transitions in calpastatin subdomains A and C. Protein Sci. 12(10), 2327–2336 (2013)
Nirenberg, M., Leder, P.: RNA codewords and protein synthesis. The effect of trinucleotides upon the binding of SRNA to ribosomes. Science 145, 1399–1407 (1964)
OMIM database. https://www.omim.org/
Ouellet, J.: RNA fluorescence with light-up aptamers. Front. Chem. 28(4), 29 (2016)
Paithankar, H., Tarang, G.S., Parvez, F., Marathe, A., Joshi, M., Chugh, J.: Inherent conformational plasticity in dsRBDs enables interaction with topologically distinct RNAs. Biophys. J. 121(6), 1038–1055 (2022)
Paloni, M., Bailly, R., Ciandrini, L., Barducci, A.: Unraveling molecular interactions in liquid-liquid phase separation of disordered proteins by atomistic simulations. J. Phys. Chem. B. 124(41), 9009–9016 (2020)
Pederson, T.: Nuclear RNA-protein interactions and messenger RNA processing. J. Cell Biol. 97, 1321–1326 (1983)
Peng, S.S., Chen, C.Y., Xu, N., Shyu, A.B.: RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17, 3461–3470 (1998)
Peng, S., Li, W., Yao, Y., Xing, W., Li, P., Chen, C.: Phase separation at the nanoscale quantified by dcFCCS. Proc. Natl. Acad. Sci. USA 117(44), 27124–27131 (2020)
Perrone-Bizzozero, N., Bolognani, F.: Role of HuD and other RNA-binding proteins in neural development and plasticity. J. Neurosci. Res. 68, 121–126 (2002)
Pichon, X., Wilson, L.A., Stoneley, M., Bastide, A., King, H.A., Somers, J., Willis, A.E.: RNA binding protein/RNA element interactions and the control of translation. Curr. Protein Pept. Sci. 13, 294–304 (2012)
Prashad, S., Gopal pp.: RNA-binding proteins in neurological development and disease. RNA Biol. 18, 972–987 (2021)
Pham, J., Keon, M., Brennan, S., Saksena, N.: Connecting RNA-modifying similarities of TDP-43, FUS, and SOD1 with MicroRNA dysregulation amidst a renewed network perspective of amyotrophic lateral sclerosis proteinopathy. Int. J. Mol. Sci. 21(10), 3464 (2020)
Rabouille, C., Alberti, S.: Cell adaptation upon stress: the emerging role of membrane-less compartments. Curr. Opin. Cell Biol. 47, 34–42 (2017)
Rahman, R., Xu, W., Jin, H., Rosbash, M.: Identification of RNA-binding protein targets with HyperTRIBE. Nat. Protoc. 13, 1829–1849 (2018)
Ramanathan, M., Majzoub, K., Rao, D.S., Neela, P.H., Zarnegar, B.J., Mondal, S., Roth, J.G., Gai, H., Kovalski, J.R., Siprashvili, Z., Palmer, T.D., Carette, J.E., Khavari, P.A.: RNA-protein interaction detection in living cells. Nat. Methods 15, 207–212 (2018)
Ramanathan, M., Porter, D.F., Khavari, P.A.: Methods to study RNA-protein interactions. Nat. Methods. 16(3), 225–234 (2019)
Ray, D., et al.: A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 (2013)
Ray, D., Kazan, H., Chan, E.T., Peña Castillo, L., Chaudhry, S., Talukder, S., Blencowe, B.J., Morris, Q., Hughes, T.R.: Rapid and systematic analysis of the RNA recognition specificities of RNAbinding proteins. Nat. Biotechnol. 27, 667–670 (2009)
RBP2GO https://rbp2go.dkfz.de/
Riback, J.A., Zhu, L., Ferrolino, M.C., Tolbert, M., Mitrea, D.M., Sanders, D.W., Wei, M.T., Kriwacki, R.W., Brangwynne, C.P.: Composition-dependent thermodynamics of intracellular phase separation. Nature 581(7807), 209–214 (2020)
Reber, S., Jutzi, D., Lindsay, H., Devoy, A., Mechtersheimer, J., Levone, B.R., Domanski, M., Bentmann, E., Dormann, D., Mühlemann, O., Barabino, S.M.L., Ruepp, M.D.: The phase separation-dependent FUS interactome reveals nuclear and cytoplasmic function of liquid-liquid phase separation. Nucleic Acids Res. 49(13), 7713–7731 (2021)
Ries, R.J., Zaccara, S., Klein, P., Olarerin-George, A., Namkoong, S., Pickering, B.F., Patil, D.P., Kwak, H., Lee, J.H., Jaffrey, S.R.: m6A enhances the phase separation potential of mRNA. Nature 7765, 424–428 (2019)
Rino, J., Desterro, J.M., Pacheco, T.R., Gadella, T.W., Jr., Carmo-Fonseca, M.: Splicing factors SF1 and U2AF associate in extra spliceosomal complexes. Mol. Cell Biol. 28, 3045–3057 (2008)
Rios, C., Warren, D., Olson, B., Abbott, A.L.: Functional analysis of microRNA pathway genes in the somatic gonad and germ cells during ovulation in C. elegans. Dev. Biol. 426(1), 115–125 (2017)
Roeder, R.G.: 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol. 26, 783–791 (2019)
Schilling, J., Broemer, M., Atanassov, I., Duernberger, Y., Vorberg, I., Dieterich, C., Dagane, A., Dittmar, G., Wanker, E., van Roon-Mom, W., Winter, J., Krauß, S.: Deregulated splicing is a major mechanism of RNA-induced toxicity in Huntington’s disease. J. Mol. Biol. 431, 1869–1877 (2019)
Shamoo, Y., Abdul-Manan, N., Williams, K.R.: Multiple RNA binding domains (RBDs) just don’t add up. Nucleic Acids Res. 23, 725–728 (1995)
Shchepachev, V., Bresson, S., Spanos, C., Petfalski, E., Fischer, L., Rappsilber, J., Tollervey, D.: Defining the RNA interactome by total RNA-associated protein purification. Mol. Syst. Biol. 15, e8689 (2019)
Simm, S., Einloft, J., Mirus, O., Schleiff, E.: 50 years of amino acid hydrophobicity scales: revisiting the capacity for peptide classification. Biol. Res. 49(1), 31 (2016)
Simon, M.D.: Capture hybridization analysis of RNA targets (CHART). Curr. Protoc. Mol. Biol. (2013). https://doi.org/10.1002/0471142727.mb2125s101
Siprashvili, Z., Webster, D.E., Kretz, M., Johnston, D., Rinn, J.L., Chang, H.Y., Khavari, P.A.: Identification of proteins binding coding and non-coding human RNAs using protein microarrays. BMC Genomics 13, 633 (2012)
Smith, T., Villanueva, E., Queiroz, R.M.L., Dawson, C.S., Elzek, M., Urdaneta, E.C., Willis, A.E., Beckmann, B.M., Krijgsveld, J., Lilley, K.S.: Organic phase separation opens up new opportunities to interrogate the RNA-binding proteome. Curr. Opin. Chem. Biol. 54, 70–75 (2020)
Soto, C., Pritzkow, S.: Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 21, 1332–1340 (2018)
Spierer, P., Zimmerman, R.A., Mackie, G.A.: RNA-protein interactions in the ribosome. Binding of 50-S-subunit proteins to 5’ and 3’ terminal segments of the 23-S RNA. Eur. J. Biochem. 52, 459–468 (1975)
Spirin, A.S.: Messenger ribonucleoproteins (informosomes) and RNA-binding proteins. Mol. Biol. Rep. 5, 53–57 (1979)
Stefl, R., Skrisovska, L., Allain, F.H.: RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Rep. 6, 33–38 (2005)
Stojković, V., Chu, T., Therizols, G., Weinberg, D.E., Fujimori, D.G.: miCLIP-MaPseq, a substrate identification approach for radical SAM RNA methylating enzymes. J. Am. Chem. Soc. 140, 7135–7143 (2018)
Takanami, M., Yan, Y., Jukes, T.H.: Studies on the site of ribosomal binding of f2 bacteriophage RNA. J. Mol. Biol. 12, 761–773 (1965)
Taylor, N.O., Wei, M.T., Stone, H.A., Brangwynne, C.P.: Quantifying dynamics in phase-separated condensates using fluorescence recovery after photobleaching. Biophys. J. 117(7), 1285–1300 (2019)
Thelen, M.P., Kye, M.J.: The role of RNA binding proteins for local mRNA translation: implications in neurological disorders. Front. Mol. Biosci. 6, 161 (2020)
Tissieres, A., Watson, J.D.: Ribonucleoprotein particles from Escherichia coli. Nature 182, 778–780 (1958)
Tome, J.M., Ozer, A., Pagano, J.M., Gheba, D., Schroth, G.P., Lis, J.T.: Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-RNA affinity profiling. Nat. Methods. 11, 683–688 (2014)
Tsai, B.P., Wang, X., Huang, L., Waterman, M.L.: Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol. Cell Proteomics 10(M110), 007385 (2011)
Ungewickell, E., Garrett, R., Ehresmann, C., Stiegler, P., Fellner, P.: An investigation of the 16-S RNA binding sites of ribosomal proteins S4, S8, S15, and S20 FROM Escherichia coli. Eur. J. Biochem. 51, 165–180 (1975)
Van Nostrand, E.L., Freese, P., Pratt, G.A., et al.: A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020)
Vanderweyde, T., Youmans, K., Liu-Yesucevitz, L., Wolozin, B.: Role of stress granules and RNA-binding proteins in neurodegeneration: a mini-review. Gerontology 59, 524–533 (2013)
Wang, X., Lu, Z., Gomez, A., Hon, G.C., Yue, Y., Han, D., Fu, Y., Parisien, M., Dai, Q., Jia, G., Ren, B., Pan, T., He, C.: N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505(7481), 117–120 (2013)
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., Gong, Z., Wang, Q., Huang, J., Tang, C., Zou, T., Yin, P.: Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534(7608), 575–578 (2016)
Wang, Y., Medvid, R., Melton, C., Jaenisch, R., Blelloch, R.: DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39(3), 380–385 (2007)
Wei, C.M., Moss, B.: Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16(8), 1672–1676 (1977)
Wells, D.G.: RNA-binding proteins: a lesson in repression. J. Neurosci. 26, 7135–7138 (2006)
West, J.A., Mito, M., Kurosaka, S., Takumi, T., Tanegashima, C., Chujo, T., Yanaka, K., Kingston, R.E., Hirose, T., Bond, C., Fox, A., Nakagawa, S.: Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 214(7), 817–830 (2016)
Xiao, W., Adhikari, S., Dahal, U., Chen, Y.S., Hao, Y.J., Sun, B.F., Sun, H.Y., Li, A., Ping, X.L., Lai, W.Y., Wang, X., Ma, H.L., Huang, C.M., Yang, Y., Huang, N., Jiang, G.B., Wang, H.L., Zhou, Q., Wang, X.J., Zhao, Y.L., Yang, Y.G.: Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 61(4), 507–519 (2016)
Youn, J.Y., Dunham, W.H., Hong, S.J., Knight, J.D.R., Bashkurov, M., Chen, G.I., Bagci, H., Rathod, B., MacLeod, G., Eng, S.W.M., Angers, S., Morris, Q., Fabian, M., Côté, J.F., Gingras, A.C.: High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell. 69(3), 517-532.e11 (2018)
Zaccara, S., Ries, R.J., Jaffrey, S.R.: Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20(10), 608–624 (2019)
Zeng, F., Peritz, T., Kannanayakal, T.J., Kilk, K., Eiríksdóttir, E., Langel, U., Eberwine, J.: A protocol for PAIR: PNA-assisted identification of RNA binding proteins in living cells. Nat. Protoc. 1, 920–927 (2006)
Zhang, X., Mahamid, J.: Addressing the challenge of in situ structural studies of RNP granules in light of emerging opportunities. Curr. Opin. Struct. Biol. 65, 149–158 (2020)
Zhang, Z., Sun, W., Shi, T., Lu, P., Zhuang, M., Liu, J.L.: Capturing RNA-protein interaction via CRUIS. Nucleic Acids Res. 48, e52 (2020)
Zheng, G., Dahl, J.A., Niu, Y., Fedorcsak, P., Huang, C.M., Li, C.J., Vågbø, C.B., Shi, Y., Wang, W.L., Song, S.H., et al.: ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49(1), 18–29 (2013)
Zheng, X., Cho, S., Moon, H., Loh, T.J., Jang, H.N., Shen, H.: Detecting RNA-protein interaction using end-labeled biotinylated RNA oligonucleotides and immunoblotting. Methods Mol. Biol. 1421, 35–44 (2016)
Acknowledgements
Work in the authors’ laboratory is supported by research funding from Indian Institute of Science Education and Research Berhampur, Department of Science and Technology-SERB. PM is a KVPY fellow. RSB is a DBT Ramalingaswami Fellow. The authors have made the best efforts to cite as many original citations and review articles as possible. However, due to the vast scope of the review topic, there may be some references which may not have been cited.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mishra, P., Sankar, S.H.H., Gosavi, N. et al. RNA nucleoprotein complexes in biological systems. Proc.Indian Natl. Sci. Acad. 88, 300–323 (2022). https://doi.org/10.1007/s43538-022-00087-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43538-022-00087-0