Abstract
We explored the most influential stand-scaled drivers of ectomycorrhizal, terricolous saprotrophic, and wood-inhabiting (main functional groups) macrofungal species richness in mixed forests by applying regression models. We tested 67 potential explanatory variables representing tree species composition, stand structure, soil and litter conditions, microclimate, landscape structure, and management history. Within the main functional groups, we formed and modeled guilds and used their drivers to more objectively interpret the drivers of the main functional groups. Terricolous saprotrophic fungi were supported by air humidity and litter mass. Ectomycorrhizal fungi were suppressed by high soil nitrogen content and high air temperature. Wood saprotrophs were enhanced by litter pH (deciduous habitats), deadwood cover, and beech proportion. Wood saprotrophic guilds were determined often by drivers with hidden effects on all wood saprotrophs: non-parasites: total deadwood cover; parasites: beech proportion; white rotters: litter pH; brown rotters: air temperature (negatively); endophytes: beech proportion; early ruderals: deciduous stands that were formerly meadows; combative invaders: deciduous tree taxa; heart rotters: coarse woody debris; late stage specialists: deciduous deadwood. Terricolous saprotrophic cord formers positively responded to litter mass. Studying the drivers of guilds simultaneously, beech was a keystone species to maintain fungal diversity in the region, and coniferous stands would be more diverse by introducing deciduous tree species. Guilds were determined by drivers different from each other underlining their different functional roles and segregated substrate preferences. Modeling guilds of fungal species with concordant response to the environment would be powerful to explore and understand the functioning of fungal communities.
Similar content being viewed by others
Introduction
To understand biodiversity, ecologists often construct statistical models to identify the drivers best able to explain species pattern–environment relationships (Guisan and Zimmermann 2000). One of the most widely used indices of biodiversity is species richness (Noss 1990), which, according to more recent studies, can be misleading as a measure of habitat quality in the evaluation of naturalness (Paillet et al. 2010) or in studying the effects of management-related habitat factors on biodiversity (Lelli et al. 2019) because it also counts the introduced species in a habitat. Furthermore, when species richness was tested in a multi-taxon study and used to describe the biodiversity of different organism groups, its relationships (congruence) were highly variable across spatial scales (Burrascano et al. 2018). Thus, species richness, as a surrogate of biodiversity, must be applied with an extra caution. However, it does have some biological value in characterizing the species diversity–environment interactions of a single organism group and provides possibility to build simple, univariate statistical models with easily comparable results.
Both phylogenetically and functionally, fungi establish highly diverse communities consuming a considerable portion of global terrestrial production as decomposers (saprotrophs), mutualists, or pathogens (biotrophs), but our knowledge of their ecology and functioning in the ecosystem is still limited (Wardle and Lindahl 2014). Previous attempts to find the most influential stand-scaled drivers of macrofungal species richness in temperate forests have focused on the main functional groups separately: wood-inhabiting, ECM, and terricolous saprotrophic fungi. Wood-inhabiting fungi have the ability to degrade the cell wall components of woody plants providing nutrients and opening habitats for other organism groups nesting in or feeding on dead wood, like several bacterial, insect, bird, and mammalian species (Stokland et al. 2012). Therefore, it was frequently reported that wood-inhabiting fungal species richness is driven by the amount, decay stage, diameter, and species identity (Abrego and Salcedo 2013) of dead wood, as well as the fragmentation, and presence or absence of bark cover and patches of epixylic vegetation (Ruokolainen et al. 2018) along with substrate-related but indirectly affecting factors like stand age (Nordén and Paltto 2001) and host tree diversity (Purhonen et al. 2020; Krah et al. 2018). The chemical characteristics (Krah et al. 2018; Boddy 2001) and the microclimatic variation (Salerni et al. 2002) within the wood were also highlighted to be important. Among the non-substrate-related factors, the significance of interspecific competition within wood-inhabiting fungal trait assemblages (Bässler et al. 2016) was revealed with the need of the spatio-temporal availability of dead wood units (Bässler et al. 2010). However, studying the effects of habitat fragmentation, the opposite was also observed (Jönsson et al. 2017). Mycorrhizal fungi, all with both biotrophic and saprotrophic activities, are vital symbionts in roots supporting plant growth by facilitating water, mineral, and nutrient uptake (Dighton 2003). They provide fungal hormones, vitamins, protection against toxic compounds and pathogens for plant individuals (Smith and Read 2008); moreover, common mycorrhizal networks for interplant communication and nutrient share resulting higher stability for plant communities (Simard et al. 2012). Playing these roles, the drivers of ECM fungi are difficult to reveal at stand scale using substrate or host-related variables. In previous studies, ECM macrofungal species richness was found to be formed by several, not substrate- or host-related factors such as season (Courty et al. 2008), dispersal limitation (Peay et al. 2007), and interspecific competition on the root surface (Van Nuland and Peay 2020; Kennedy 2010). Substrate- and host-related drivers were also found to be significant underlining the importance of the low nitrogen content (Cox et al. 2010), pH, temperature and moisture content of soil (Smith and Read 2008), and the species identity of the host plant (Kernaghan et al. 2003). Terricolous saprotrophic fungi, never forming mycorrhiza but colonizing litter and buried plant debris, are responsible for the complete breakdown of plant biopolymers, maintaining the carbon and nutrient cycles in the ecosystem and making these compounds accessible for mycorrhizal fungi, insects, and bacteria (Berg and McClaugherty 2014). Less information is available on the determinants of terricolous saprotrophic fungal species richness, but the factors reported to have influential effects include: litter quantity and pH, phosphorus and carbon levels in the soil (Reverchon et al. 2010; Tyler 1991), and air temperature (McMullan-Fisher et al. 2009). Within these three main functional groups, the stand-scaled drivers forming the species richness of guilds have, to our knowledge, never been studied purposefully. However, Ohtonen et al. (1997) have already underlined the need for the exploration of the drivers of fungal guilds to better understand the fundamental roles of macrofungi in nature.
Within species richness models, the biological interpretation of drivers is sometimes problematic: both the studied environment and the species composition of the modeled species group must be considered. Reviewing the environmental aspects, the relative influence of drivers depends on the scale of the investigation (Lilleskov and Parrent 2007); and regularly, different drivers emerge with significant effects along ecological (e.g., moisture or elevation; Sundqvist et al. 2013) and geographical (Bahram et al. 2018) gradients. Moreover, the importance of drivers always varies among habitats due to environmental heterogeneity (Stein et al. 2014), and those drivers that actually limit fungal growth can have disproportionately high influence (Jumpponen and Egerton-Warburton 2005). Focusing on the modeled species group, there is a potential source of misleading results when a group of functionally highly different species is tested instead of a functionally homogeneous species group. When modeling a functionally highly heterogeneous species group, there is a strong possibility to fail to detect concordant (statistically strong and clear) species response to the environment because each fungal species has different environmental requirements (Mori et al. 2016).
Working with functionally homogeneous species groups, models could reveal highly significant but scarcely interpretable drivers. Such drivers often have indirect effects on the studied species group. In this case, one can only hypothesize a suitable biological interpretation, which is a general problem in the evaluation of results in ecological modeling. To overcome this problem, we completed a strategic hierarchical subset of the three main macrofungal functional groups, created a nested data structure, modeled each subset separately with the same methods, and evaluated the drivers of subsets simultaneously to obtain additional evidence for an objective interpretation of the drivers of the main functional groups.
We aimed to (i) explore the stand-scaled drivers of the species richness of wood-inhabiting, ECM, and terricolous saprotrophic macrofungi (main functional groups) in temperate forests, West Hungary and (ii) provide evidence that these groups involve guilds shaped by different environmental drivers. Many of these remain hidden when only the main functional groups are modeled.
Materials and methods
Study site
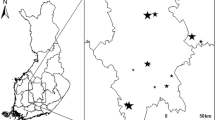
We conducted mycological surveys in Őrség National Park (ŐNP, 440 km2), West Hungary (46° 51′–55′ N, 16° 06′–24′ E (Fig. 1A). ŐNP is defined by the annual precipitation range of 700–800 mm and situated at the border of the subalpine and Pannonian climate zones. The mean annual temperature range is 9.1–9.8 °C, and the mean minimum and maximum temperatures are − 7.4 to 6.0 °C in winter and 13.5–23.8 °C in summer (Hungarian Meteorological Service). Nutrient-poor brown forest soils (planosols or luvisols) are the most frequent soil types (Halász et al. 2006) with a topsoil pH range of 4.0–4.8 (Juhász et al. 2011). ŐNP is a perfect place to examine the effects of different tree taxa on forest-dwelling macrofungi due to its highly mixed forest stands with the dominance of beech (Fagus sylvatica), sessile and pedunculate oak (Quercus petraea and Q. robur), hornbeam (Carpinus betulus), and Scots pine (Pinus sylvestris) (Tímár et al. 2002). In our previous study (Kutszegi et al. 2015), we provided a comprehensive description of the region considering its climate, soil characteristics, tree species and historical land uses.
Environmental data collection
To reduce the effects of edaphic heterogeneity on macrofungal species richness, we chose 35 mature, 70–100-year-old, monodominant, and mixed stands of beech, oak, Scots pine and hornbeam of varying proportions, which were located in relatively flat areas and not influenced directly by surface waters. Within each stand, we assigned a plot of 30 m × 30 m for macrofungal surveys (Fig. 1B). See Siller et al. (2013) for GPS coordinates of plots. The smallest distance among plots is 500 m to minimize the interfering effects of spatial autocorrelation. The plots cover a low elevation range (250–350 m a.s.l.) to reduce the effects of an underlying elevation gradient on fungal species richness.
We measured 67 potential environmental variables representing tree species composition, stand structure, soil and litter conditions, microclimate, landscape structure, and management history to test for relationships with macrofungal species richness (Table 1). Field measurements of environmental variables are detailed in Appendix A and in Kutszegi et al. (2015).
Fungal data
Due to the large total area (31,500 m2) of sampling units, we completed sporocarp surveys instead of DNA sequence-based identification techniques to characterize macrofungal species richness. We sampled basidiomycetes (excluding most of the resupinate non-poroid taxa) and ascomycetes that develop sporocarps larger than 2 mm. We focused exclusively on species that belong to the main fungal functional groups: ECM, terricolous saprotrophic, and wood-inhabiting fungi (descriptions in Table S1). To sample the fungal species fruiting in spring, summer, or in autumn, we organized three sampling visits: in August 2009 (for 8 days), in May 2010 (8 days) and during September–November 2010 (due to the perfect fruiting conditions, for 48 days). We fixed a species list in each plot, in each sampling visit, and merged the results of the three visits to obtain the total observed species richness of plots. We detailed our species identification and nomenclatural procedures in Kutszegi et al. (2015) and Siller et al. (2013).
Statistical analysis
We divided our raw fungal species richness data into hierarchical subsets (guilds) based on the ecological functioning (F; 14 subsets) and host tree preference (H; 11 subsets) of fungal species (Fig. 2c, Tables S1, S2). Literature about the life strategy (functioning) of macrofungal species is mainly restricted to wood-inhabiting fungi; consequently, this classification is completed almost exclusively for this fungal group. The life strategy of several ECM and terricolous saprotrophic fungi is still unknown (Dighton 2003); therefore, we almost failed to separate guilds within them. Forming guilds, we classified the wood-inhabiting species according to their pathogenicity, type of rot, and life strategy following Pirttilä and Frank (2011) and Boddy et al. (2008).
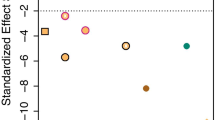
Comparison of R2-values of 26 quasi-Poisson linear models shown in Fig. 3; grouping of models by main functional groups (a), division methods (b), and classification levels within divisions (c). Part (c) details briefly the hierarchical arrangement of models. We detected slight dissimilarities but no evidences of significant (p < 0.01) differences among R2-values. We performed ANOVA and Student’s t tests to complete multiple (a, c) and paired (b, c) comparisons, respectively (see text for p values). ECM fungi had consequently lower R2-values (a). Models on subsets separated based on fungal host tree preferences were stronger because we included explanatory variables that characterize the species composition of trees (b). Probably due to concordant species responses, models done on functional groups and on species groups by host tree preference tended to be a bit stronger toward lower classification levels (c). Box metrics: central line, median; box, inter-quartile range; whisker, 1.5 × inter-quartile range
For the exploration of the relationships between species richness data and environmental variables, we built multiple regression models, applied general linear modeling (GLM) based on Faraway (2005, 2006), and used the statistical software R for Windows 3.0.1 (R Core Team 2013).
Prior to building GLMs:
-
(a)
We checked the normality of each potential explanatory and response variables. Some explanatory variables were needed to be ln-transformed to satisfy the criterion of normality (Table 1). For the response variables (Table S1), no logarithmic transformations were needed.
-
(b)
We centered and standardized the explanatory variables by standard deviation.
-
(c)
We corrected each of our response variables for sampling bias because our field survey in autumn lasted for 48 days until the end of (or beyond) the fruiting period of some fungal species. We numbered the days of this sampling period from 1 to 48 to create a “sampling time” variable and then applied partial correlations according to Legendre and Legendre (1998) to remove (partial out) the distorting effects of sampling bias.
-
(d)
We screened the explanatory variables for collinearity. First, calculating correlation matrices, we completed correlations pairwise between each response and each potential explanatory variable to select the explanatory variables with a homogeneous relationship stronger than |r| = 0.35 to a response variable. Among these explanatory variables, we screened the ones with a stronger collinearity of |r| > 0.45 and excluded the variable(s) with a weaker effect to the response variable.
Due to the under-dispersion of variances, pinpointed by the R package “AER” (Kleiber and Zeileis 2008), we fitted quasi-Poisson GLMs to our species data following Zuur et al. (2009). We applied a manual backward selection for the full models by dropping the explanatory variables stepwise until all the remaining variables met the criterion of p < 0.05 within the models (based on deviance analyses with F-statistics). We calculated a pseudo-R2 to our quasi-Poisson GLMs based on McFadden (1974), and applied deviance analyses with F-statistics to establish a statistical reliability for the final models. For each final model, we checked the validation graphs to assess the homogeneity and normality of residuals, homoscedasticity, and Cook statistics to reveal whether any observation has extreme values of the explanatory variables. Finally, using the R package “faraway” (Faraway 2002), we calculated a variance inflation factor (Faraway 2005) for each explanatory variable included in the final models to screen them for multicollinearity.
Applying ANOVA or Student’s t test, we compared the R2 values of models done on the subsets of (i) the three main functional groups, (ii) our two groups of models (subsets by ecological functioning vs. host tree preference), and (iii) the classification levels within the two model groups of point (ii). To help understand the drivers of the three main functional groups, we classified the drivers of their subsets based on their significance and the number of subsets they form. By this simultaneous evaluation of models, we got the possibility to highlight those drivers of subsets that (i) cannot be detected by the exclusive modeling of the main functional groups and (ii) possibly have indirect effects on the main functional groups.
Results
Macrofungal diversity
We observed 676 fungal taxa (Table S2), obtaining 4067 records. Of these, we collected 896, 274, and 2897 records in August 2009, in May 2010, and during autumn 2010, respectively.
Different guilds, different drivers: substrate and host were decisive
We separated and modeled 25 subsets of the full sample (Fig. 3); hierarchical arrangements of subsets are in Fig. 2c, more statistical details of models in Figs. S1 and S2.
Species richness models (quasi-Poisson GLMs) on the environmental drivers of macrofungal functional groups. To reveal the drivers cannot be detected by the exclusive test of the full sample, we divided the full sample based on the ecological function (F1–F14) and host tree preference (H1–H11) of fungal species and built a separate model on each subset (guild). The simultaneous examination of the drivers of subsets helps understand the drivers of the main (F1–F3) functional groups. A: total number of modeled taxa, B: drivers with negative (blue) and positive (red) effects, C: percent proportion of variance explained, D: significance codes: *** < 0.0001; ** < 0.001; * < 0.01; (″) < 0.05; (′) < 0.1. Serial numbers of models in bold; response variables by capital letters. Hierarchical arrangements of subsets are in Fig. 2c; more statistical details in Figs. S1 and S2
The environmental variables revealed by all of our models can be arranged into our six variable groups with the following proportions: soil and litter conditions (28%; 23 significant variables out of the 80 revealed in total), tree species composition (25%), stand structure (24%), microclimate (15%), landscape structure (4%), and management history (4%). In general, the three main functional groups (models F1–F3) and other large subsets with more than 200 modeled species (F5, F7) were driven principally by the general environmental requirements of macrofungi: microclimate (temperature) and substrate properties (pH of litter, soil nitrogen content, and amount of dead wood). By contrast, the small subsets of 7–49 species were mainly formed by more specific drivers representing the stand structure, the chemical components of the soil and litter layer, and the surrounding landscape.
Specifically, the species richness of terricolous saprotrophic fungi (F1) was driven by air humidity and litter mass. ECM fungi (F2) were negatively determined by the nitrogen content of soil and air temperature. Wood-inhabiting macrofungal species richness (F3) was principally formed by litter pH, dead wood cover, and beech proportion.
Wood-inhabiting fungal guilds (F5–F14) were mainly determined by well-interpretable drivers not detected at the level of all wood-inhabiting fungi. However, two drivers (litter pH and beech proportion) influenced both the group of all wood-inhabiting fungi and some of its subsets, but usually with highly different importance. Non-parasites (F5) were principally shaped by total dead wood cover and litter pH, whereas beech proportion was the most important driver for parasites (F6). White rotters (F7) were found to be a substrate-related guild following stands with high litter pH and high proportion of deciduous dead wood. By contrast, brown rotters (F8) were mainly driven by microclimate (air temperature, negatively), while the effects of host (beech) and substrate (total volume of coniferous logs and snags) were less pronounced. Wood-inhabiting fungal guilds defined by life strategies (F9–F14) and describing consecutive stages of wood decay were determined by drivers different from each other. Endophytes, with the ability to switch from a biotrophic to a saprotrophic lifestyle, favored stands with high beech proportion. Early ruderals, colonizing freshly exposed dead wood units, preferred forest stands that were formerly meadows and had a relatively high litter pH. Combative invaders were selective to stands with a high total proportion of deciduous tree taxa, less surrounded by forests, and with higher litter pH. Heart rotters responded significantly to higher CWD volumes. Late stage specialists were determined exclusively by the total volume of deciduous logs and snags. Wood-inhabiting cord formers avoided stands with a high moss cover on the forest floor.
Terricolous saprotrophic cord formers (F4) were positively affected by litter mass, while FWD mass proportion and minimum air temperature had negative effects.
Guilds formed by host tree preference (H1–H11), expectedly, responded most significantly to variables describing tree species composition (deciduous vs. coniferous). However, the other drivers within the models carry valuable information. All the models on fungal species selective for coniferous habitats (H2, H7, and H8) pointed out a specific requirement of higher host tree diversity in conifer-dominated stands. Host generalists (H3, H9–H11) responded to similar drivers, but with different importance as that of the full sample and the main functional groups (F1–F3). Contrasting model F1 with H9 about terricolous saprotrophic fungi, the two models revealed the same results: high air humidity and high litter mass were needed. Host generalist ECM fungi (H10) mainly required higher tree species richness with higher moss cover; the low soil nitrogen content had only marginal effects. By contrast, all ECM fungi (F2) were primarily shaped by soil nitrogen content. Host generalist wood-inhabiting fungi (H11) were determined by dead wood cover followed by more beech and less Scots pine, whereas all wood-inhabiting fungi (F3) responded primarily to litter pH and only secondarily to dead wood cover and beech proportion.
Indirect effects
We classified the drivers revealed by the GLMs in Fig. 3 to separate the most important (determinative) ones from those of with marginal or probable indirect effects (Table 2). Checking the drivers for common occurrences in the same models, we marked (asterisk) three drivers in category A to have probable indirect effects on macrofungi in the region.
All wood-inhabiting fungi (F3) were most significantly driven by litter pH. To try to explain this unexpected result, we compared the drivers of all the 11 subsets that were significantly formed by litter pH. In eight out of 11 cases (F3, F5–F7, F11, H1, H5, and H6), we observed that litter pH was selected together with drivers characterizing deciduous habitats. In addition, model H1, about all fungal species selective for deciduous habitats, emphasized litter pH with the strongest effect.
Despite the fact that we checked our explanatory variables for collinearity within the models, we did find some cases when two drivers repeatedly co-occurred in the same model. For example, moss cover was very likely to co-occur with beech proportion; see model F3, F5–F7, and F14 (in 5 models out of 7). Also, Scots pine proportion showed the same pattern with beech in all models in which it was significant (F3, F5, and H11).
Comparison of the explanatory powers of models
We compared the R2-values of our GLMs and found slight, non-significant differences (Fig. 2): among the main functional groups (ANOVA, p = 0.2679, a), division methods (Student’s t test, p = 0.2267, b), and among classification levels within division methods (for functional groups, ANOVA, p = 0.519; for host tree preference, Student’s t test, p = 0.6703, c).
Discussion
Drivers of macrofungal functional groups and guilds
The species richness of macrofungal communities in the ŐNP was mainly formed by drivers describing soil and litter conditions, tree species composition, and stand structure (in this order). Microclimate, landscape structure, and management history had minor effects. Similar results have long been reported by several authors (Daws et al. 2020; Krah et al. 2018; Boddy et al. 2008; Smith and Read 2008; Kernaghan et al. 2003) concluding that macrofungi, in general, are primarily substrate and host-restricted organisms.
Terricolous saprotrophic fungi were positively determined by air humidity and litter mass. McMullan-Fisher et al. (2009) did not directly measure air humidity but found that a higher annual rainfall and air temperature are important for all macrofungi in various (in both dry and humid) Eucalyptus forests of Tasmania. Tyler (1991) revealed that the experimentally doubled amount of litter enhances the sporocarp production of soil saprotrophs which is in line with our results. It is interesting that, in our study, microclimate had stronger effects on terricolous saprotrophs, which are thought to be a principally substrate-restricted organism group in temperate forests (Boddy et al. 2008).
ECM fungi were negatively driven by soil nitrogen content and air temperature. This result is concomitant with the results of other authors, e.g., Cox et al. (2010) and Peter et al. (2001) who have reported that the high soil nitrogen content has generally negative effects on the fruiting of several ECM fungal species in temperate forests. However, the opposite was also confirmed in severely nitrogen-limited boreal forests (Perez-Moreno and Read 2001). In our study, the general preference of ECM fungal species to lower air temperatures cannot be explained clearly based on the drivers of our guilds. The phenomenon may be clarified by the fact that the studied region is a home to many ice age relic ECM fungal species with boreal distribution centers, for which the area is a refuge; see Siller et al. (2013) and Vasas and Locsmándi (1995) for species lists. Several boreal ECM species are selective to lower temperatures (Smith and Read 2008).
Wood-inhabiting fungi, surprisingly, was determined by litter pH. Dead wood cover and beech proportion were less significant, while the total moss cover (together on soil and dead wood units) and Scots pine proportion had marginal (and probably indirect) effects. To our knowledge, litter pH also has indirect effects on wood saprotrophs and highlights a general preference of deciduous habitats (detailed in the next chapter). It can be explained by the fact that most (92%) of our collected wood saprotrophs was a white rotter, generally preferring angiosperm wood (Krah et al. 2018), and two-third of all (63%) were exactly selective to deciduous trees. Wood-inhabiting fungi are substrate-restricted organisms (Boddy et al. 2008); thus, their high dependence on available dead wood is obvious and was repeatedly reported by many authors: Heilmann-Clausen et al. (2014), Stokland et al. (2012), and Kirk and Cowling (1984). The strong effect of beech in our study on wood-inhabiting fungi can be explained by both the quick production of large dead wood volumes in the fast-growing beech stands (Heine et al. 2019) and the relatively high cellulose content of beech wood (Schwarze and Baum 2000). Beech forests were mentioned among the richest habitats in wood saprotrophs (Küffer and Senn-Irlet 2005); and according to Heine et al. (2019), beech specifically promotes the diversity and species richness of wood-inhabiting fungi in Europe.
Cord-forming litter saprotrophs were enhanced by litter mass and suppressed by fine woody debris proportion. Litter is the main substrate of this guild which was well-illustrated by Boddy et al. (2009) demonstrating that cord formers mainly grow at the soil–litter interface interconnecting and consuming litter components. Large amounts of FWD in litter may inhibit hyphal growth by facilitating the aeration and slowing the compaction of the lower litter layer making the environment less favorable for fungi (Berg and McClaugherty 2014).
Non-parasitic and parasitic (necrotrophic) wood-inhabiting fungi showed highly differing requirements. Most species were shared between non-parasites and all wood-inhabiting fungi; therefore, both groups were shaped by similar drivers with fine differences in importance. By contrast, necrotrophic parasites were clearly determined by beech proportion. According to Schwarze et al. (2000), beech harbors a relatively rich community of parasitic fungi in Central European temperate forests. This phenomenon is not completely understood but can be explained by the wide geographical distribution, large body size, fast growth, and thin, vulnerable bark (Biggs 1992) of beech, as well as its high cellulose and low lignin and tannin contents (Schwarze and Baum 2000). In our data set, half of the tree parasites indeed preferred beech as a primary host.
White rotters and brown rotters differed strongly in their drivers. We mainly found white rotters in deciduous habitats, which is in accord with the strong, positive effects of litter pH, deciduous dead wood, and beech proportion in their model. Their requirements were very similar to those revealed for all wood-inhabiting fungi. They were found to be substrate-restricted. Based on the phylogeny of more than 1000 wood-inhabiting fungal species from the Northern Hemisphere, Krah et al. (2018) found that most white rot fungi are angiosperm specialists owing to the closely simultaneous diversification of these fungal group and angiosperms in the Cretaceous. In contrast, brown rotters were primarily suppressed by high air temperatures and enhanced by high amounts of coniferous dead wood. We failed to join this temperature relation to previous studies, but we attempt to explain the phenomenon with the generally cooler microclimate of coniferous stands compared to that of deciduous ones. Heat loss is generally higher in coniferous stands where the canopy openness is high due to the narrow leaves. According to Krah et al. (2018) and Hibbett and Donoghue (2001), most brown rot fungi are gymnosperm specialists supposedly because their lineages originated during the diversification of gymnosperms.
Similar to parasitic wood-inhabiting fungi, endophytes were linked strongly to high beech proportion. This is consistent with the results of Unterseher and Schnittler (2010) and Sieber (2007) who mentioned beech as one of the most species rich host for endophytes and other wood saprotrophs (Heine et al. 2019) in Central European forests. Here, beech plays a similar role in the ŐNP significantly forming the species richness of several subsets. The chemical and structural characteristics of beech wood are detailed above for parasites; these properties probably also explain the strong effects of beech on endophytes.
Early ruderals were driven primarily by forest management history; they responded to the high proportion of meadows surrounding the sampling unit c.a. 160 years ago. Caution must be used in interpretation of this result because their model has little explanatory power and a very low number of tested species. Moreover, all of our sampling units were assigned in stands older than 70 years which is enough time for some dead wood to be produced and many early ruderals (even some late stage specialists) to establish (Boddy and Heilmann-Clausen 2008; Boddy 2001). Once some dead wood is produced, the habitat is open for decomposer fungi; therefore, it is unimaginable that a variable expressing a 160-year-old situation could significantly affect this group.
Combative invaders were determined by the high proportion of deciduous trees and substrates. These combative secondary colonizers can cause rapid white rot on fallen logs and branches (Boddy and Heilmann-Clausen 2008) dominating the decomposition of angiosperm wood (Hibbett and Donoghue 2001). Interestingly, they were negatively affected by the proportion of forests in the landscape which can be likely explained by the general low amount of dead wood in the surrounding old stands close to their age of harvest. Following silvicultural activities, cutting areas are opened and some substrates (mainly stamps) can be produced for these fungi.
Heart-rot agents responded significantly to CWD volume. This is in accord with the conclusions of Moore et al. (2008) about the relatively high nitrogen requirement of heart rotters among wood decay fungi. To acquire enough nitrogen for the development of big, perennial sporocarps, heart rotters must establish large mycelial domains and colonize large dying trees and dead wood units.
The species richness of late stage specialists was significantly higher in stands with high amounts of deciduous dead wood. Surprisingly, the proportion of decayed logs had no detectable effects probably due to the general low amounts of this substrate in our sampling units. Toward the final stage of wood decomposition, Renvall (1995) found an increasing number of white rotters at the expense of brown rotters because white rotters have enzymes to degrade the more complex wood components (lignin and hemicellulose), in which the well decayed dead wood units are generally richer.
Wood-inhabiting cord formers were negatively influenced by moss cover. It is known that soil respiration and decomposition rates can be significantly reduced by a well-developed moss layer (Nilsson and Wardle 2005). Studying Armillaria mellea rhizomorphs growing in deeper soil layers, Morrison (1976) reported decreasing oxygen and increasing carbon-dioxide levels to have limiting effects on rhizomorph growth and wood decomposition. Modeling the species composition of all wood-inhabiting fungi on logs of deciduous trees, Ruokolainen et al. (2018) and Heilmann-Clausen et al. (2005) also observed moss cover with significant effects and concluded that it may have stabilizing effects on the microclimatic environment within the wood and can cause shifts in the species composition rather than in the species richness of wood-inhabiting fungi.
Drivers affecting indirectly
The group of all wood-inhabiting fungi was primarily determined by litter pH. To our knowledge, litter pH has never been detected as a driver of this functional group. Here, in 8 models of 11, litter pH was significant and selected together with drivers characterizing deciduous habitats. This provides some evidence that litter pH rather represents a general preference of deciduous habitats not the special impacts of litter pH per se. Another variable of probable indirect effects on wood-inhabiting fungi was moss cover, always with suppressing effects and acting as a follower of beech proportion (in 5 models out of 7). Working in shaded, beech-dominated habitats with a generally poor moss layer, Heilmann-Clausen and Christensen (2005) also confirmed this observation. Similarly, Scots pine proportion was always co-acted with beech proportion and rather had marginal effects on wood-inhabiting fungi. We found no drivers with probable indirect effects for terricolous saprotrophic and ECM fungi.
The importance of tree diversity and tree species richness was specifically revealed in coniferous habitats exclusively forming those guilds that species were selective for coniferous hosts. Conifer-dominated forests harbor a distinctive mycota with species of specific abilities to decompose needle litter and coniferous wood (Humphrey et al. 2000). In such habitats, increasing tree diversity by broadleaved tree species can make major contributions to a richer fungal community.
Is it worth modeling fungal guilds?
Former studies on environmental drivers have mainly focused on all the wood-inhabiting (e.g., Krah et al. 2018; Ruokolainen et al. 2018; Abrego and Salcedo 2013), ECM (e.g., Van Nuland and Peay 2020; Cox et al. 2010; Smith and Read 2008), or terricolous saprotrophic fungi (e.g., Reverchon et al. 2010; Boddy et al. 2009; McMullan-Fisher et al. 2009) in the studied region. Within these groups, guilds have rarely been formed and modeled (but see Štursová et al. 2020; Heilmann-Clausen et al. 2014, and Boddy 2001) due to the limited knowledge on the life strategies, substrate preferences, and environmental requirements of several fungal species (Dighton 2003). Working with guilds is difficult because many fungal species can switch among life strategies along changing environmental conditions (Zanne et al. 2020). Hence, it is impossible to establish discrete boundaries among them (Pirttilä and Frank 2011). Moreover, numerous different grouping methods can be applied onto the same species pool. Guilds can be defined as community components with similar effects on, or with concordant responses to the environment (Hooper et al. 2002). Guilds can also be defined as groups of species competing for the same resources, inhabiting a certain microhabitat, or community components exploiting different resources but in related ways (Simberloff and Dayan 1991). Here, we also set up guilds based on different point of views. We tried to overcome the aforementioned problems by constructing guilds according to criteria that are related fundamentally to the general environmental drivers of macrofungi, representing substrate quality and quantity, life history, and host tree taxonomy. In whatever aspects we formed guilds, we often got groups with less than 40 species, for which we also obtained models with strong (R2 × 100 > 50) explanatory powers. Moreover, for many guilds, we usually revealed drivers that were not detected (were hidden) modeling the entire functional group containing the guild.
Proofs of concordant species response
A species richness model will be strong when: (i) the tested species group actually forms a guild with concordant species responses to the environment, (ii) exactly those environmental variables were measured that are truly important for the species group, and (iii) the model contains several (3–5) significant variables. The positive effect of concordant species response on the explanatory power of models can also be observed in our results: models done on host generalists often detected weaker responses compared to the models on host specialists.
Our generally weaker models on ECM fungi can be explained by the following reasons: (i) we probably did not measure those environmental variables that really influence these species in the region, (ii) it is not yet clear which of these species form a guild and have similar environmental requirements, and (iii) it is highly probable that our model on all ECM fungi contains several guilds (species) that are driven principally by different environmental drivers. Deveautour et al. (2020) also suggested numerous guilds within ECM communities. It would be especially worthwhile for ECM fungi to find the environmental requirements of single species using linear regression models. With such models, we could define ECM guilds that consist of species with similar responses to the environment and thus better understand the functioning of the whole ECM community.
Our model on all litter saprotrophs was weaker than the model on cord-forming litter saprotrophs. Due to this, several embedded litter saprotrophic guilds can be suggested within the main functional group of litter saprotrophs. Long-term studying litter decomposition, Štursová et al. (2020) revealed consecutive successional phases along the breakdown process to be dominated by different litter saprotrophic fungal guilds.
Limitations
We exclusively sampled sporocarps for a short duration (2 yrs) of field visits to measure macrofungal species richness. It is an underestimate of the total species richness in the studied region because we consistently skipped to register microfungi (those that never produce sporocarps) and certainly have overlooked some of those macrofungal taxa that produce inconspicuous sporocarps (Tóth and Barta 2010). Also, sometimes decades of field surveys are needed to register most of the less frequently fruiting macrofungal species (Straatsma et al. 2001) due to the high variation among years in the sporocarp production of species (Fernández-Toirán et al. 2006). It must be also noted that our total species pool of 676 species exceeds the usual sample size of inventories with a similar sampling strategy and intensity. It can be explained by the very humid weather in autumn 2010, when 71% of all records were obtained including many rare species (Siller et al. 2013). The life strategies of several fungal species are still unknown; therefore, our functional classification, even for the represented guilds, is incomplete.
Conclusions
In the ŐNP, the stand-scaled species richness of all wood-inhabiting, ECM, and terricolous saprotrophic macrofungi were principally driven by substrate properties (litter pH, soil nitrogen content, amount of dead wood) and microclimate (air humidity). Of these drivers, litter pH had indirect effects on wood-inhabiting fungi, representing deciduous habitats, while the others were generally well-interpretable.
Within these main groups, we formed guilds and modeled them with the same methods. The guilds were conditioned by more specific drivers characterizing the key elements of fungal diversity: host tree species richness, dead wood diameter and quality, and landscape properties. Several drivers of guilds were with undetectable (hidden) effects on the main functional groups. The simultaneous evaluation of drivers of guilds helped to interpret the indirectly affecting drivers of the main functional groups in an evidence-based way.
Due to limited literature data, it was not yet possible to form complete guilds within ECM and litter saprotrophic fungi according to species life history strategies; however, the presence of embedded guilds within these main groups was highly likely after comparing the explanatory power of models.
It would be worthwhile for all macrofungi to find the environmental requirements of each species using linear regression models or composing and modeling fungal guilds with species of similar substrate preferences or exploitation strategies. With such models, we could define fungal community components that consist of species with similar responses to the environment to better understand the fundamental roles of macrofungi in ecosystem functioning.
Abbreviations
- AL:
-
Ammonium lactate
- ANOVA:
-
Analysis of variance
- CWD:
-
Coarse woody debris
- DBH:
-
Diameter at breast height
- DW:
-
Dead wood
- ECM:
-
Ectomycorrhizal
- FWD:
-
Fine woody debris
- GLM:
-
Generalized linear model
- ŐNP:
-
Őrség National Park
References
Abrego, N., & Salcedo, I. (2013). Variety of woody debris as the factor influencing wood-inhabiting fungal richness and assemblages: Is it a question of quantity or quality? Forest Ecology and Management, 291, 377–385. https://doi.org/10.1016/j.foreco.2012.11.025.
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature, 560, 233–237. https://doi.org/10.1038/s41586-018-0386-6.
Bässler, C., Müller, J., Cadotte, M. W., Heibl, C., Bradtka, J. H., Thorn, S., et al. (2016). Functional response of lignicolous fungal guilds to bark beetle deforestation. Ecological Indicators, 65, 149–160. https://doi.org/10.1016/j.ecolind.2015.07.008.
Bässler, C., Müller, J., Dziock, F., & Brandl, R. (2010). Effects of resource availability and climate on the diversity of wood-decaying fungi. Journal of Ecology, 98, 822–832. https://doi.org/10.1111/j.1365-2745.2010.01669.x.
Berg, B., & McClaugherty, C. (2014). Plant litter: Decomposition, humus formation, carbon sequestration (3rd ed., p. 315). Berlin: Springer-Verlag.
Biggs, A. R. (1992). Anatomical and physiological responses of bark tissues to mechanical injury. In R. A. Blanchette & A. R. Biggs (Eds.), Defense mechanisms of woody plants against fungi (pp. 13–40). Berlin: Springer-Verlag.
Boddy, L. (2001). Fungal community ecology and wood decomposition processes in angiosperms: From standing tree to complete decay of coarse woody debris. Ecological Bulletins, 49, 43–56.
Boddy, L., Frankland, J. C., & van West, P. (Eds.). (2008). Ecology of Saprotrophic Basidiomycetes (p. 372). London: The British Mycological Society, Academic Press.
Boddy, L., & Heilmann-Clausen, J. (2008). Basidiomycete community development in temperate angiosperm wood. In L. Boddy, J. C. Frankland, & P. van West (Eds.), Ecology of saprotrophic Basidiomycetes (pp. 211–237). London: The British Mycological Society, Academic Press.
Boddy, L., Hynes, J., Bebber, D. P., & Fricker, M. D. (2009). Saprotrophic cord systems: Dispersal mechanisms in space and time. Mycoscience, 50, 9–19. https://doi.org/10.1007/s10267-008-0450-4.
Burrascano, S. R., de Andrade, B., Paillet, Y., Ódor, P., Antonini, G., Bouget, C., et al. (2018). Congruence across taxa and spatial scales: Are we asking too much of species data? Global Ecology and Biogeography, 27, 980–990. https://doi.org/10.1111/geb.12766.
Courty, P.-E., Franc, A., Pierrat, J.-C., & Garbaye, J. (2008). Temporal changes in the ectomycorrhizal community in two soil horizons of a temperate oak forest. Applied and Environment Microbiology, 74, 5792–5801. https://doi.org/10.1128/AEM.01592-08.
Cox, F., Barsoum, N., Lilleskov, E. A., & Bidartondo, M. I. (2010). Nitrogen availability is a primary determinant of conifer mycorrhizas across complex environmental gradients. Ecology Letters, 13, 1103–1113. https://doi.org/10.1111/j.1461-0248.2010.01494.x.
Daws, S. C., Cline, L. A., Rotenberry, J., Sadowsky, M. J., Staley, C., Dalzell, B., et al. (2020). Do shared traits create the same fates? Examining the link between morphological type and the biogeography of fungal and bacterial communities. Fungal Ecol., 46, 100948. https://doi.org/10.1016/j.funeco.2020.100948.
Deveautour, C., Chieppa, J., Nielsen, U. N., Boer, M. M., Mitchell, C., Horn, S., et al. (2020). Biogeography of arbuscular mycorrhizal fungal spore traits along an aridity gradient, and responses to experimental rainfall manipulation. Fungal Ecology, 46, 100899. https://doi.org/10.1016/j.funeco.2019.100899.
Dighton, J. (2003). Fungi in ecosystem processes (p. 432). New York: Marcel Dekker Inc.
Faraway, J. J. (2002). Practical regression and ANOVA using R (pp. 117–120). Unpublished manuscript. https://cran.r-project.org/doc/contrib/Faraway-PRA.pdf. Retrieved from, May 5, 2016.
Faraway, J. J. (2005). Linear models with R (p. 229). Boca Raton: Texts in Statistical Science, Chapman & Hall/CRC.
Faraway, J. J. (2006). Extending the linear model with R: Generalized linear, mixed effects and nonparametric regression models (p. 301). Boca Raton: Taylor & Francis Group, LLC.
Fernández-Toirán, L. M., Ágreda, T., & Olano, J. M. (2006). Stand age and sampling year effect on the fungal fruit body community in Pinus pinaster forests in central Spain. Canadian Journal of Botany, 84, 1249–1258. https://doi.org/10.1139/B06-087.
Guisan, A., & Zimmermann, N. E. (2000). Predictive habitat distribution models in ecology. Ecological Modelling, 135, 147–186. https://doi.org/10.1016/S0304-3800(00)00354-9.
Halász, G., Bartha, D., Bidló, A., Berki, I., Király, G., Koloszár, J., et al. (2006). Magyarország erdészeti tájai (Forest regions of Hungary) (p. 154). Budapest: Magyar Állami Erdészeti Szolgálat.
Heilmann-Clausen, J., Aude, E., & Christensen, M. (2005). Cryptogam communities on decaying deciduous wood—Does tree species diversity matter? Biodiversity and Conservation, 14, 2061–2078. https://doi.org/10.1007/s10531-004-4284-x.
Heilmann-Clausen, J., Aude, E., van Dort, K., Christensen, M., Piltaver, A., Veerkamp, M., et al. (2014). Communities of wood-inhabiting bryophytes and fungi on dead beech logs in Europe—Reflecting substrate quality or shaped by climate and forest conditions? Journal of Biogeography, 41, 2269–2282. https://doi.org/10.1111/jbi.12388.
Heilmann-Clausen, J., & Christensen, M. (2005). Wood-inhabiting macrofungi in Danish beech-forests—Conflicting diversity patterns and their implications in a conservation perspective. Biological Conservation, 122, 633–642. https://doi.org/10.1016/j.biocon.2004.10.001.
Heine, P., Hausen, J., Ottermanns, R., Schäffer, A., & Roß-Nickoll, M. (2019). Forest conversion from Norway spruce to European beech increases species richness and functional structure of aboveground macrofungal communities. Forest Ecology and Management, 432, 522–533. https://doi.org/10.1016/j.foreco.2018.09.012.
Hibbett, D. S., & Donoghue, M. J. (2001). Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in Homobasidiomycetes. Systematic Biology, 50, 215–242. https://doi.org/10.1080/10635150121079.
Hooper, D. U., Solan, M., Symstad, A., Diaz, S., Gessner, M. O., Buchmann, N., et al. (2002). Species diversity, functional diversity, and ecosystem functioning. In M. Loreau, S. Naeem, & P. Inchausti (Eds.), Biodiversity and ecosystem functioning: Synthesis and perspectives (pp. 195–208). New York: Oxford University Press.
Humphrey, J. W., Newton, A. C., Peace, A. J., & Holden, E. (2000). The importance of conifer plantations in northern Britain as a habitat for native fungi. Biological Conservation, 96, 241–252. https://doi.org/10.1016/S0006-3207(00)00077-X.
Jönsson, M. T., Ruete, A., Kellner, O., Gunnarsson, U., & Snäll, T. (2017). Will forest conservation areas protect functionally important diversity of fungi and lichens over time? Biodiversity and Conservation, 26, 2547–2567. https://doi.org/10.1007/s10531-015-1035-0.
Juhász, P., Bidló, A., Ódor, P., Heil, B., & Kovács, G. (2011). Őrségi erdőtalajok széntartalmi vizsgálata (Investigation of soil carbon content in Őrség, West Hungary). Talajvédelem (Suppl.), Szeged, 377–382.
Jumpponen, A., & Egerton-Warburton, L. M. (2005). Mycorrhizal fungi in successional environments: A community assembly model incorporating host plant, environmental, and biotic filters. In J. Dighton Jr., J. F. White Jr., & P. Oudemans (Eds.), The fungal community: Its organization and role in the ecosystem (3rd ed., pp. 139–168). Boca Raton: Taylor & Francis Group.
Kennedy, P. (2010). Ectomycorrhizal fungi and interspecific competition: Species interactions, community structure, coexistence mechanisms, and future research directions. New Phytologist, 187, 895–910. https://doi.org/10.1111/j.1469-8137.2010.03399.x.
Kernaghan, G., Widden, P., Bergeron, Y., Légaré, S., & Paré, D. (2003). Biotic and abiotic factors affecting ectomycorrhizal diversity in boreal mixed-woods. Oikos, 102, 497–504. https://doi.org/10.1034/j.1600-0706.2003.12415.x.
Kirk, T. K., & Cowling, E. B. (1984). Biological decomposition of solid wood. In R. M. Rowell (Ed.), The chemistry of solid wood (pp. 455–487). Washington, DC: American Chemical Society.
Kleiber, Ch., & Zeileis, A. (2008). Applied econometrics with R (p. 221). New York: Springer.
Krah, F.-S., Bässler, C., Heibl, C., Soghigian, J., Schaefer, H., & Hibbett, D. S. (2018). Evolutionary dynamics of host specialization in wood-decay fungi. BMC Evolutionary Biology, 18, 119. https://doi.org/10.1186/s12862-018-1229-7.
Küffer, N., & Senn-Irlet, B. (2005). Influence of forest management on the species richness and composition of wood-inhabiting basidiomycetes in Swiss forests. Biodiversity and Conservation, 14, 2419–2435. https://doi.org/10.1007/s10531-004-0151-z.
Kutszegi, G., Siller, I., Dima, B., Takács, K., Merényi, Zs, Varga, T., et al. (2015). Drivers of macrofungal species composition in temperate forests, West Hungary: Functional groups compared. Fungal Ecol., 17, 69–83. https://doi.org/10.1016/j.funeco.2015.05.009.
Legendre, P., & Legendre, L. (1998). Numerical ecology (2nd ed., p. 853). Amsterdam: Elsevier Science B.V.
Lelli, C., Bruun, H. H., Chiarucci, A., Donati, D., Frascaroli, F., Fritz, Ö., et al. (2019). Biodiversity response to forest structure and management: Comparing species richness, conservation relevant species and functional diversity as metrics in forest conservation. Forest Ecology and Management, 432, 707–717. https://doi.org/10.1016/j.foreco.2018.09.057.
Lilleskov, E. A., & Parrent, J. L. (2007). Can we develop general predictive models of mycorrhizal fungal community–environment relationships? New Phytologist, 174, 250–256. https://doi.org/10.1111/j.1469-8137.2007.02023.x.
McFadden, D. (1974). Conditional logit analysis of qualitative choice behavior. In P. Zarembka (Ed.), Frontiers in econometrics: Economic theory and mathematical economics (pp. 105–142). New York: Academic Press.
McMullan-Fisher, S. J. M., Kirkpatrick, J. B., May, T. W., & Pharo, E. J. (2009). Surrogates for macrofungi and mosses in reservation planning. Conservation Biology, 24, 730–736. https://doi.org/10.1111/j.1523-1739.2009.01378.x.
Moore, D., Gange, A. C., Gange, E. G., & Boddy, L. (2008). Fruit bodies: Their production and development in relation to environment. In L. Boddy, J. C. Frankland, & P. van West (Eds.), Ecology of saprotrophic Basidiomycetes (pp. 79–103). London: The British Mycological Society, Academic Press.
Mori, A. S., Isbell, F., Fujii, S., Makoto, K., Matsuoka, S., & Osono, T. (2016). Low multifunctional redundancy of soil fungal diversity at multiple scales. Ecology Letters, 19, 249–259. https://doi.org/10.1111/ele.12560.
Morrison, D. J. (1976). Vertical distribution of Armillaria mellea rhizomorphs in soil. Transactions of the British Mycological Society, 66, 393–399. https://doi.org/10.1016/S0007-1536(76)80207-0.
Nilsson, M.-C., & Wardle, D. A. (2005). Understory vegetation as a forest ecosystem driver: Evidence from the northern Swedish boreal forest. Frontiers in Ecology and the Environment, 3, 421–428. https://doi.org/10.1890/1540-9295(2005)003%5b0421:uvaafe%5d2.0.co;2.
Nordén, B., & Paltto, H. (2001). Wood-decay fungi in hazel wood: Species richness correlated to stand age and dead wood features. Biological Conservation, 101, 1–8. https://doi.org/10.1016/S0006-3207(01)00049-0.
Noss, R. F. (1990). Indicators for monitoring biodiversity: A hierarchical approach. Conservation Biology, 4, 355–364. https://doi.org/10.1111/j.1523-1739.1990.tb00309.x.
Ohtonen, R., Aikio, S., & Väre, H. (1997). Ecological theories in soil biology. Soil Biology & Biochemistry, 11, 1613–1619. https://doi.org/10.1016/S0038-0717(97)00063-1.
Paillet, Y., Bergès, L., Hjältén, J., Ódor, P., Avon, C., Bernhardt-Römermann, M., et al. (2010). Biodiversity differences between managed and unmanaged forests: Meta-analysis of species richness in Europe. Conservation Biology, 24, 101–112. https://doi.org/10.1111/j.1523-1739.2009.01399.x.
Peay, K. G., Bruns, T. D., Kennedy, P. G., Bergemann, S. E., & Garbelotto, M. (2007). A strong species–area relationship for eukaryotic soil microbes: Island size matters for ectomycorrhizal fungi. Ecology Letters, 10, 470–480. https://doi.org/10.1111/j.1461-0248.2007.01035.x.
Perez-Moreno, J., & Read, D. J. (2001). Exploitation of pollen by mycorrhizal mycelial systems with special reference to nutrient recycling in boreal forests. Proceedings of the Royal Society of London, Series B: Biological Sciences, 268, 1329–1335. https://doi.org/10.1098/rspb.2001.1681.
Peter, M., Ayer, F., & Egli, S. (2001). Nitrogen addition in a Norway spruce stand altered macromycete sporocarp production and below-ground ectomycorrhizal species composition. New Phytologist, 149, 311–325. https://doi.org/10.1046/j.1469-8137.2001.00030.x.
Pirttilä, A. M., & Frank, A. C. (Eds.). (2011). Endophytes of forest trees: Biology and applications (p. 319). Dordrecht: Springer.
Purhonen, J., Ovaskainen, O., Halme, P., Komonen, A., Huhtinen, S., Kotiranta, H., et al. (2020). Morphological traits predict host-tree specialization in wood-inhabiting fungal communities. Fungal Ecology, 46, 100863. https://doi.org/10.1016/j.funeco.2019.08.007.
R Core Team. (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Retrieved from, http://www.R-project.org. Accessed 26 Apr 2013.
Renvall, P. (1995). Community structure and dynamics of wood-rotting Basidiomycetes on decomposing conifer trunks in northern Finland. Karstenia, 35, 1–51. https://doi.org/10.29203/ka.1995.309.
Reverchon, F., del Ortega-Larrocea, P. M., & Pérez-Moreno, J. (2010). Saprophytic fungal communities change in diversity and species composition across a volcanic soil chronosequence at Sierra del Chichinautzin, Mexico. Annals of Microbiology, 60, 217–226. https://doi.org/10.1007/s13213-010-0030-7.
Ruokolainen, A., Shorohova, E., Penttilä, R., Kotkova, V., & Kushnevskaya, H. (2018). A continuum of dead wood with various habitat elements maintains the diversity of wood-inhabiting fungi in an old-growth boreal forest. European Journal of Forest Research, 137, 707–718. https://doi.org/10.1007/s10342-018-1135-y.
Salerni, E., Laganà, A., Perini, C., Loppi, S., & De Domonicus, V. (2002). Effects of temperature and rainfall on fruiting of macrofungi in oak forests of the Mediterranean area. The Israel Journal of Plant Sciences, 50, 189–198. https://doi.org/10.1560/GV8J-VPKL-UV98-WVU1.
Schwarze, F. W. M. R., & Baum, S. (2000). Mechanisms of reaction zone penetration by decay fungi in wood of beech (Fagus sylvatica). New Phytologist, 146, 129–140. https://doi.org/10.1046/j.1469-8137.2000.00624.x.
Schwarze, F. W. M. R., Engels, J., & Mattheck, C. (2000). Fungal strategies of wood decay in trees (p. 185). Berlin: Springer-Verlag.
Sieber, T. N. (2007). Endophytic fungi in forest trees: Are they mutualists? Fungal Biology Reviews, 21, 75–89. https://doi.org/10.1016/j.fbr.2007.05.004.
Siller, I., Kutszegi, G., Takács, K., Varga, T., Merényi, Zs, Turcsányi, G., et al. (2013). Sixty-one macrofungi species new to Hungary in Őrség National Park. Mycosphere, 4, 871–924. https://doi.org/10.5943/mycosphere/4/5/3.
Simard, S. W., Beiler, K. J., Bingham, M. A., Deslippe, J. R., Philip, L. J., & Teste, F. P. (2012). Mycorrhizal networks: Mechanisms, ecology and modelling. Fungal Biology Reviews, 26, 39–60. https://doi.org/10.1016/j.fbr.2012.01.001.
Simberloff, D., & Dayan, T. (1991). The guild concept and the structure of ecological communities. Annual Review of Ecology, Evolution, and Systematics, 22, 115–143. https://doi.org/10.1146/annurev.es.22.110191.000555.
Smith, S. E., & Read, D. J. (2008). Mycorrhizal symbiosis (3rd ed., p. 787). Oxford: Elsevier Ltd.
Stein, A., Gerstner, K., & Kreft, H. (2014). Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecology Letters, 17, 866–880. https://doi.org/10.1111/ele.12277.
Stokland, J. N., Siitonen, J., & Jonsson, B. G. (2012). Biodiversity in dead wood (p. 509). New York: Cambridge University Press.
Straatsma, G., Ayer, F., & Egli, S. (2001). Species richness, abundance, and phenology of fungal fruit bodies over 21 years in a Swiss forest plot. Mycological Research, 105, 515–523. https://doi.org/10.1017/S0953756201004154.
Štursová, M., Šnajdr, J., Koukol, O., Tláskal, V., Cajthaml, T., & Baldrian, P. (2020). Long-term decomposition of litter in the montane forest and the definition of fungal traits in the successional space. Fungal Ecology, 46, 100913. https://doi.org/10.1016/j.funeco.2020.100913.
Sundqvist, M. K., Sanders, N. J., & Wardle, D. A. (2013). Community and ecosystem responses to elevational gradients: Processes, mechanisms, and insights for global change. Annual Review of Ecology, Evolution, and Systematics, 44, 261–280. https://doi.org/10.1146/annurev-ecolsys-110512-135750.
Tímár, G., Ódor, P., & Bodonczi, L. (2002). The characteristics of forest vegetation of the Őrség Landscape Protected Area. Kanitzia, 10, 109–136.
Tóth, B. B., & Barta, Z. (2010). Ecological studies of ectomycorrhizal fungi: An analysis of survey methods. Fungal Diversity, 45, 3–19. https://doi.org/10.1007/s13225-010-0052-2.
Tyler, G. (1991). Effects of litter treatments on the sporophore production of beech forest macrofungi. Mycological Research, 95, 1137–1139. https://doi.org/10.1016/S0953-7562(09)80561-3.
Unterseher, M., & Schnittler, M. (2010). Species richness analysis and ITS rDNA phylogeny revealed the majority of cultivable foliar endophytes from beech (Fagus sylvatica). Fungal Ecology, 3, 366–378. https://doi.org/10.1016/j.funeco.2010.03.001.
Van Nuland, M. E., & Peay, K. G. (2020). Symbiotic niche mapping reveals functional specialization by two ectomycorrhizal fungi that expands the host plant niche. Fungal Ecology, 46, 100960. https://doi.org/10.1016/j.funeco.2020.100960.
Vasas, G., & Locsmándi, Cs. (1995). The macroscopic fungi (Basidiomycetes) of Őrség, Western Hungary. Savaria, 22(2), 265–294.
Wardle, D. A., & Lindahl, B. D. (2014). Disentangling global soil fungal diversity. Science, 346, 1052–1053. https://doi.org/10.1126/science.aaa1185.
Zanne, A. E., Powell, J. R., Flores-Moreno, H., Kiers, E. T., van ‘t Padje, A., & Cornwell, W. (2020). Finding fungal ecological strategies: Is recycling an option? Fungal Ecology, 46, 100902. https://doi.org/10.1016/j.funeco.2019.100902.
Zuur, A. F., Ieno, E. N., Walker, N. J., Saveliev, A. A., & Smith, G. M. (2009). Mixed effects models and extensions in ecology with R. Statistics for biology and health (p. 574). New York: Springer.
Acknowledgements
This study was supported by the Hungarian Scientific Research Fund (OTKA, K79158), the National Research, Development and Innovation Office of Hungary (GINOP 2.3.3-15-2016-00019), and the Directorate of Őrség National Park. Authors are indebted to Bence Kovács, Attila Lengyel, and two anonymous reviewers for their valuable comments on the manuscript.
Funding
Open access funding provided by University of Veterinary Medicine.
Author information
Authors and Affiliations
Contributions
GK wrote and edited the first draft of the manuscript, conducted statistical analyses, prepared figures, and provided the methodology. IS, BD, GK, KT, ZsM, TV, and GT collected fungal data and performed identifications. AB completed soil and litter data. PÓ organized the study and provided continuous participation in data analyses and writing. All authors contributed substantially to revisions.
Corresponding author
Additional information
Nomenclature: MycoBank (www.mycobank.org, accessed 19–20 April 2019).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kutszegi, G., Siller, I., Dima, B. et al. Revealing hidden drivers of macrofungal species richness by analyzing fungal guilds in temperate forests, West Hungary. COMMUNITY ECOLOGY 22, 13–28 (2021). https://doi.org/10.1007/s42974-020-00031-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42974-020-00031-6