Abstract
Coronavirus (Covid) is a severe acute respiratory syndrome infectious disease, spreads primarily between human beings during close contact, most often through the coughing, sneezing, and speaking small droplets. A retrospective surveillance research is conducted in India during 30th January–21st March 2020 to gain insight into Covid’s epidemiology and spatial distribution. Voronoi statistics is used to draw attention of the affected states from a series of polygons. Spatial patterns of disease clustering are analyzed using global spatial autocorrelation techniques. Local spatial autocorrelation has also been observed using statistical methods from Getis-Ord G *i . The findings showed that disease clusters existed in the area of research. Most of the clusters are concentrated in the central and western states of India, while the north-eastern countries are still predominantly low-rate of clusters. This simulation technique helps public health professionals to identify risk areas for disease and take decisions in real time to control this viral disease.
Similar content being viewed by others
1 Introduction
A New Coronavirus-nCoV’ was identified in December 2019 and subsequently renamed SARSCoV-2 in Wuhan, Hubei, China, resulting in extreme acute respiratory syndromes [1]. Covid is a pandemic that is actively expanding throughout the world and a unique challenge for the community’s healthcare, economy and lifestyle. Countries are grappling with many tactics in order to minimize the spread of Covid: ban collection, close schools, stop transportation, lock towns, enforce curfew, and seal places, but unable to contain it effectively [2]. The time is required to locate the risk assessment on a site basis to take prompt preventive measures. Globally, there are 48,93,195 cases of coronavirus, while the death toll is 3,22,861. Taking into account the updates of the Ministry of Health on Wednesday (May 20, 2020), India received a total of 1,06,750 COVID cases which include 61,149 active cases, 42,298 cure and 3303 deaths [3]. In the last 24 h, there have been 5611 new cases and 140 deaths. The rate of recovery is 39.62%.
A key element of epidemiologic research, the geographical distribution of the disease, is demonstrated by the importance given to the “person, place and time” descriptor of health events in the classical epidemiology textbooks [4]. Geographical information systems (GIS) have today revolutionized these spaces-which, in simple terms, give the ability to view spatial or geographic information in a meaningful way, be it interactive maps or other infographics. There are numerous uncertainties in the Covid pandemic, many of them have a spatial component that contributes to the epidemic being interpreted as geographical and technically mappable [5]. In the battle against Covid in India, there have been limited Covid risk maps and application of Covid spatial epidemiology [6]. For these purposes, with the emergence of Covid as a global pandemic, the use of geospatial and statistical methods has become extremely important. Statistical modelling and spatial epidemiology in small areas have been developed in order to solve problems where disease clusters and hotspots are located. Some of the principal spatial techniques explored by Robertson [7] are spatial autocorrelation, spatial time interactions, hotspots and clusters, used throughout emerging infectious disease research.
Advances in geostatistical methods have provided for substantially improved efficiency in the processing and analysis of complex georeferenced data with multiple variables on different geographical scales, providing epidemiologists with new instruments to incorporate space and place in their study [8, 9]. The public health authorities use the disease prevalence maps as a guide to monitor and prevention programs to classify areas of excess and their possible causes (e.g. exposures to the environment or socio-demographic factors). Three major hurdles lie in interpreting and analyzing the choropleth maps: (2) visual bias due to including health data within administrative units of large varies sizes and types and (3) spatial support mismatch for the occurrence of disease and explanatory variable data that prevents direct use of correlational research, (1) extremely unsafe rates which usually occur for sparsely populated regions and/or less frequently observed Covid.
Geostatistical algorithms have been developed to filter local small-scale variations on cluster maps that enhance regional trends on a larger scale [10, 11]. Their computer requirements and the underlying assumptions about spatial patterns and distribution of risk values differ greatly in these methods. An important exploratory technique in scientific inquiry continues to be cluster analysis [12]. Spatial cluster detection depends on the geography of the activities which requires the correct and meaningful treatment of space and spatial relations, combined with the location and event attributes observed. It has to date involved the use of specific structuring and accounting methods and techniques for the distance, the outskirts, the contiguity, geographical irregularity and so on. Understanding the distribution of Covid cases in India with the use of geostatistical analysis approach, will help inform Covid control programme at smaller scale. The specific objectives are to use of spatial auto-correlation technique to analyze Covid spatial pattern and identify clusters with statistically significant hotspots of the disease. Present study focused on basic geostatistical approaches to deal with Covid clustering pattern in India.
2 Data collection and processing
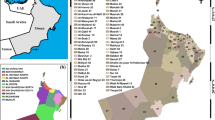
The incorporated in this study includes all information reported by Government of India up to the latest of 21st May 2020 [3]. This report considers all states and Union Territories as well (Fig. 1). In this report, the confirmed Covid cases and death due to Covid along with the transmission types is considered. The skewness of the Covid incidence data is measured in terms of the third moment of the mean of the distribution. If the distribution is symmetric, the skewness is zero. The kurtosis describes the extent of peak of the distribution, measured by fourth moment of the mean. The distributions with kurtosis lower than 3 are known as platykurtic.
Voronoi polygons are created so that every location within a state is closer to the affected state than any other non-affected states. After the polygons are created, neighbours of affected states are defined as any other sample locations whose boundary shares a border with the chosen affected states. The Voronoi map tool provides a number of local statistics (mean, median, entropy, inter-quartile range) in which polygons can be assigned or calculated [13]. The status value is the average value that the states and their neighbour’s states calculate which is used for further processing and analysis.
3 Methods
3.1 Directional distribution of disease pattern
Directional distribution, namely the standard deviation ellipse (SDE), was used each year to calculate the directional pattern and to provide compactness and orientation information on the dispersion of the infected Covid. The standard distance measurement in x- and y-directions is a common way of measuring the pattern for a certain group of areas [14]. Each of these measures describes an ellipse axis that covers the distribution of characteristics [15]. The SDE determines the default x-coordinates and y-coordinates of the centre in order to determine the Ellipse’s axes. The ellipse enables one to see if the distribution of features is elongated and therefore has a particular orientation. For both the infected Covid, we have used a standard deviation which account for approximately 68% of all input variables [16]. In order to compare the spatial patterns of the Covid infected and local source, a series of additional measurements and data including an axial ratio and coordinates of each ellipse were collected for six days interval.
3.2 Covid-2019 spatial pattern
The ArcGIS 10.0 statistical toolkit for global autocorrelation (Moran’s I) and Getis-Ord G *i were used in the identification of statistically significant Covid clusters for the various states in India. The statistical technique of the Moran’s I evaluates the spatial autocorrelation of Covid cases in geographical areas clusters where the value of Moran’s I close to zero means that the illness is spatially random; a good value suggests spatial clusters [17]. A statistically significant estimation of Moran’s I (p < 0.05, z score ~ 1.96) suggests that neighbouring districts have similar Covid cases under the null-hypothesis that Covid distribution on a regional scale is absolutely spatially random to determine whether the spatial trend is clustered, dispersed or random. The optimized high-low cluster produces using the Getis-Ord G *i statistics of statistically relevant cluster (e.g., states with high Covid cases). Spatial outliers comprise high-low (a high value in a low-value states) and low–high outliers (a low-value value in a high-value states). Getis-Ord G *i value less than 1 demonstrates positive space autocorrelation whereas it suggests a value greater than 1 point to negative spatial autocorrelation [18].
To identify the spatial relationship, fixed distance band is used in which each state boundary is analysed within the context of neighbouring state boundary. Neighbouring state boundary outside the specified critical distance receive the number of cases and extent the influence on computation for the affected/non-affected states. Neighbouring state boundaries outside the critical distance receive a weight of zero and have no influence on the affected state computation. This method measures a z-score and P-values that are statistical tests to demonstrate whether a null hypothesis can be rejected or not. For statistically significant positive z-scores, the larger the z-score is, the more intense the clustering of high Covid affected states (hotspot). For statistically significant negative z-scores, the smaller the z-scores is, the more intense the clustering of low Covid incidences (cold spot). Moreover, very high positive or negative z-score are associated with the very small P-values which indicate it is unlikely that the observed spatial pattern reflects the theoretical random pattern represented by the null hypothesis. A confidence level of 99 percent we have selected which indicated that we are unwilling to reject the null hypothesis unless the probability that the pattern was created by random chance is less than a 1 percent probability.
3.3 Risk zone identification using areal interpolation
Areal interpolation is a wide variety of methods that can measure the cumulative attribute of a unit system (in this case, newly created polygons), based on the attribute data structure of another, spatially incongruous structure (in this case, the original polygons). The initial units for which the characteristic is defined are also called source units and for which the characteristic has to be measured, target units are called objective units [19, 20]. The proposed pycnophylactic areal interpolation algorithms [21] are based on different assumptions regarding the underlying distribution of Covid-2019 cases, relies only on the databeing estimated. The lattice spacing is defined to estimate the point covariances of Covid incidence, each state is overlaid with a square lattice, and a point is assigned to each intersection in the lattice. The number of cases in the source units is spatially redirected to the target units using as a weight the area that each source unit contributes to the target area [22]. It is an improvement over the area-weighting method because it does not assume a homogeneous distribution of cases, and the continuous surface eliminates sharp transitions in Covid case estimates across state boundaries.
3.4 Predicting accuracy
The predictive accuracy is measured using the root mean square error (RMSE) based on the interpolated value \({\mathop{\text{y }}^{\smile }}_{\rm j}\) in 5-km distance as follows:
Where, j represents the number of intersection units whose number of units is J. In this case, the space autocorrelation distance between two models represents an effect. Our findings show that the spatial autocorrelation in the field is necessary to take into account.
4 Results
4.1 Descriptive analysis of Covid distribution
All statistical data are processed using Microsoft Excel version 11.0. Continuous variables are conveyed as the mean, standard deviations or medians, skewness, kurtosis and range as appropriate (Table 1). Results showed mean value of Covid cases are dramatically increased from 30th January, 2020 to 21st January, 2020. In January, the mean incidence of Covid cases calculated as 0.03; whereas, on 21st May, 2020, the estimated average Covid incidence was 836.84. High skewed of Covid distribution was observed from 30th January, 2020 to 16th March, 2020. The skewness of the dataset varies between 1.50 on 22nd March, 2020 and 5.83 on 30th January, 2020. All data were positively skewed, indicated that the size of the right tail is larger than the left tail. Kurtosis is associated with the back, shoulder and peakedness of the distribution. The value of kurtosis ranged between 4.44 and 35.03. The positive value indicated platykurtic distribution. Sample size-weighted measures are beneficial since we expect a survey variable to better represent that of the population as the sample size increased. To account for this, measurements from large samples are given a higher weight than those from smaller samples.
4.2 Directional distribution pattern of Covid
The SDE method establishes a new pattern with an intermediate centre elliptic polygon for all states. Such output ellipse polygons have Covid incidence value of two normal lengths (long and short axes); ellipse orientation (Table 2). The direction is the rotation of the long clockwise axis from noon. During the period between 10th March 2020 and 09th April 2020, 27th April 2020 and 09th May, 2020 two ellipses have been generated. Figure 2 shows the series of directional distributions of the Covid infected patients in each week (from 30th January, 2020 to 21st May, 2020). Plotting ellipses for Covid outbreak during the study period may provide insight of disease spread which may be useful for in deploying mitigation strategies. The districts are spatially regular (so they are the mostly concentrated in the middle and become increasingly dense towards the periphery).
Their shapes are dissimilar from one week to another; the ellipse is generally oriented along the west–east direction. From 30th January 2020 to 16th March 2020, both the long and short axes became larger and oriented towards west–east, indicated most of the cases are found in west and eastern states of India and concentrated dispersedly within the states (Fig. 2). Since 10th March, 2020 a small ellipse are oriented towards north–south direction, and both the long and short axes are smaller which means that the standard deviations of the ellipses were decreasing and Covid cases were distributed in some north and southern states (Fig. 2c). On 16th March, 2020, the north–south SDE were shifted towards eastern direction (Fig. 2d). on 22nd March, 2020 three SDE were generated having an orientation towards south-west to north-east direction with large axes, north-east to south-west direction with small axes and a very small axes of east–east-south to north–north-west direction. This indicates that Covid cases were distributed in most the states in India and mostly concentrated in the north and north-west direction. Since 28th March to 04th April, 2020, two SDEs were generated with south-west to north-east and south-east to north-west direction. However, the size of south-east to north-west orientation ellipse is comparative smaller which indicated maximum concentration of the disease in this particular state. From 6th April, 2020 to 15th April 2020, results showed most of the cases were concentrated in the central and southern states of India. Since 15th April 2020 (Fig. 2m–o), the SDE are shifted towards west and south and oriented towards south-west to north-east, indicated that the number maximum concentration of cases is higher in this region.
4.3 Spatial autocorrelation of covid distribution
The Global Moran’s I method is an inferential statistic, meaning that the study’ findings are often interpreted in accordance with its null hypothesis. The null hypothesis of the Global Moran’s I statistics suggests that the analyzed attribute is randomly distributed between the states. Two special cases of the general cross-product data measuring spatial auto-correlation. Table 3 shows a description of the findings of the spatial autocorrelation data calculated through Moran’s I and Getis-Ord Gi* statistics on weekly infected Covid. There were statistically relevant findings from global Moran’s I test (z scores above 1.96) and suggest spatial heterogeneity. Also, statistically important were the global Covid autocorrelation figures between 30th January 2020 and 21st May 2020. Results show that the change in the Covid distribution’s spatial autocorrelation with intervals between 0.326 and 0.662 was relatively unstable. A positive value of Moran’s I suggests positive spatial autocorrelation which means a combination of high values and low values. The largest Moran’s I value indicated the strongest spatial autocorrelation of Covid affected states. In this study, the distance band was 10 km and the spatial clusters were further studied. In the absence of the norm of the data, an exponent for the transformation of the data into a normal distribution was defined in the Box-Cox transformation. The autocorrelation among the states were lower from 30th January to 10th March, 2020. Since 16th March to 15th May, higher value of Moran’s I were calculated with high Z-score value and significant P value at 99% confidence level; however, the estimated value of Moran’s I is less on 21st May 2020. The transformed data can also remove the effect on the spatial cluster analysis of extreme values.
The Getis-Ord Gi* tool evaluates each Covid infected state and compares the local situation with the global situation in the neighbouring states. The value derived through Getis-Ord Gi* statistics, z-score and P-value are represented in Table 3. These results indicate that there was positive spatial auto-correlation. Results showed all the z-score values were significant at α < 0.01 level. Hence, we could reject the null hypothesis. The spatial distribution in the data set of high and/or low values of Covid was spatially clustered more than expected if the underlying spatial processes were changed. The higher value of Getis-Ord Gi* and z-score was calculated for 16th March, 2020 and from 15th April, 2020 to 05th May, 2020 (Table 3). The calculated values of Getis-Ord Gi* from 09th May to 21st May, 2020 represented in Table 3. The lowest value of Getis-Ord Gi* observed from 30th January to 04th March, 2020.
4.4 Covid risk zone identification
Our primary aim in this analysis was the detection of Covid hotspots using statistical methods for potential intervention. By considering number of Covid cases for each state and the average measurement, a prediction surface was produced for the value of the Gaussian variable at all states in the data domain. In this interpolation the empirical co-variances were adjusted within the 90% confidence interval. Exponential model appears to be fit the data very well; most of the covariances fall within the confidence intervals. The searching neighbourhood of the predicted value of 0.0006119 was fixed for the fifth-grade obesity rates along with the smoothing factor of 0.2. The details of the model parameters (lattice spacing, lag size, mean, major range, RMSE, and average standard error) were illustrated in Table 4. The lattice spacing of the model was varied between 1.6417 and 1.8808 during the study period. The lattice spacing parameter specifies the horizontal and vertical distance between each central location of the infected states. The partial sill was increased with the change of time. This indicated that the predicted Covid incidence value of any state at that location had about 33 percent change of being obese. The mean value of the study Covid model was varied between − 0.0027 and 14.3895. The negative mean value was calculated for 30th January, 2020, 10th March–28th March, 2020 and 21st April, 2020. The estimated root mean square was varied between 0.7996 and 7.289 during the study period. This indicates that ideally good prediction of surface as it was close to ‘1’. The value of Root Mean Square Error (RMSE) was gradually increased in the study period, except on 15th May, 2020 (7.289). The average standard error in the study area was gradually increased, except on 09th April, 2020.
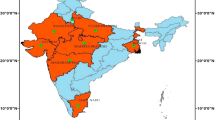
Figure 3 portrays the spatial distribution of areal interpolation of Covid cases during the period between 30th January, 2020 and 21st May 2020 for the re-aggregation of Covid cases to downscale or upscale within the state boundary. The areal interpolation tool predicts the average value of Gaussian distribution (with prediction standard error) for the affected states. Results of the analysis showed on or before 30th January, 2020 only southern states of India are affected by Covid and on 04th March 2020, very high risk (VHR) Covid zones were identified in the extreme north and central north of India and small pockets of high-riskzones are found in Telengana, Kerala state and north-west of India. On 10th March, 2020, VHR zones were identified central and south-west of India and small pockets are observed in Uttar Pradesh and Delhi. The high-risk (HR) Covid zones were identified in the entire north of India and surrounding of very high-risk (VHR) zone. On 16th March, entire central, south and south-west of India were identified as very risk Covid zone. Moreover, small pockets of VHR areas were also identified in the west and Uttar Pradesh state. The HR Covid zones were identified in the north-west and northern part of India. On 22nd March, most of the central and western states of India were falls under the VHR zone; and the north and adjacent to VHR zone. The entire north-east states are demarcated as low risk for Covid. On 28th March, 2020, the VHR areas were mostly concentrated in the central and southern states of India. The west and northern states are recognized as HR for Covid. On 3rd April, the south and central part of India were identified as VHR and HR zone were demarcated in the west and central north of India. From 09th April –15th April, 2020, Covid VHR were mostly concentrated in the central and west of the nation and HR areas were shifted towards east and disseminated on the north and south of the India. Since 21st April to 03rd May, 2020, the VHR zones were identified in the central and south-west of India. HR zone were extended towards the entire eastern zone of India. Since 03rd May 2020, the pattern is almost similar; however, low risk areas of Covid were identified in the Chhattisgarh and Odisha states. Moreover, the HR zones were extended towards entire south and extreme north-west of India.
5 Discussion
India is now the fourth most confirmed in Asia and on 19th May 2020, the number of cases exceeding the 100,000 mark is still in its fourth. The fatality rate of India is relatively low by 3.09% compared to 6.63% on 20th May 2020, worldwide (https://ourworldindata.org/-coronavirus-india?country=IND). In India, the number of persons who were present from 16 February to 29 March dropped sharply in retail and recreational areas compared with traffic from 3rd January to 6th February. On 22nd March the Government of India decided to lock 82 districts in 22 States, where confirmation of the cases had been reported until 31st March, in 22 States and Union territories. With the end of the lockdown approaching, some states proposed that the lockdown be prolonged. On 14th April, the national lockdown was extended until 3rd May, with Indian Prime Minister, Mr. Narendra Modi being able to provide relaxation from 20th April on the spacious regions. Testing began on 15th March for community transmission. A random monitoring of people with flu-like symptoms and samples of patient without any travel or associations of people infected 65 laboratories of the Health Research and the Indian Medical Research Council (DHR-ICMR) has been conducted. In 36 regions of 15 states with positive SARI patients (Severe Acute Respiratory Illnesses) were registered, the Indian Council of Medical Research (ICMR) was recommended priority confinement in patients with Severe Acute Respiratory Infection (SARI). As on 26 May, 3126,119 samples were tested and 145,380 people were confirmed positive according to the ICMR [23].
Few previous ecological studies have been conducted in India concerning Covid problems and their links to risk factors. However, very limited number of studies was conducted on Covidspatio-temporal pattern in India. The present study of India’s leading health issues’ spatial clustering could help to understand the spatial epidemiology of Covid for the implementation of regional prevention and control strategies by health departments. Traditional statistical analyses with no displays or graphics restrict their usefulness, particularly to advice policymakers on priority issues. Data are encoded as graphics or images and can be displayed more intuitively. Therefore, our study is focused on the visualization of the Covid distribution created by GIS and geostatistical technique. Spatial autocorrelation analysis and spatial cluster analysis were carried out to identify spatio-temporal pattern of Covid clusters.
The use of infectious disease of spatial analytics techniques is not new [24,25,26]. For cluster analysis of regional health issues, spatial autocorrelation measurement is useful [27, 28]. The positive values of Moran’s I and Getis-Ord G *i statistics indicate spatial autocorrelation as cluster (clustering of similar neighbour states). A significant positive spatial autocorrelation was observed in our analysis and it is corroborated with earlier results [29]. The neighbours of each state boundary were established by the edge contiguity, which gives weights to neighbours with edge sharing, and to all the others. In view of the z scores: 5.43 and 8.33 for January and April respectively, suggest that the observed clustered trend is likely to have a probability of less than 1 percent. A complete randomness hypothesis is rejected, as there are high Covid cases in neighbouring places in an urban and peri-urban area. This indicates that Covid cases are spatially clustered throughout metropolitan area and peri-urban areas. Identifying these hotspot Covid areas may be taken care and supervise regional Covid prevention programmes. This will help policymakers analyse spatial risk factors to figure out the way to move forward of health care strategies for health services preparation and implementation.
Areal interpolation has been demonstrated to be a promising method for defining endemic disease clusters [30,31,32,33].The projection of areal interpolation moves the complicated structure from high dimensional regions into lower dimensional clusters, which is essential to cluster endemic disease areas based on the neighbourhood relations. The integration of local interpolation and GIS is designed successfully to produce dynamic visualisation, which in turn helps public health officials to decide Covid management in a timely manner. Most of the infection are concentrated in the central and southern states of India. Spatial distribution pattern of Covid cases are significantly clustered and identified in the north-central and extreme north of India. Hotspot areas are mainly distributed in Maharashtra, Telengana, Karnataka, Gujrat, Madhya Pradesh and Rajasthan and cold spots areas are distributed in the north-east of India.In a future, spatial autocorrelation analysis help to understand the temporal dependence.
There are still some limitations of this analysis. In the present study, we do not include migratory population and others socio-economic parameters. In order to assess the associations between Covid and its potential factors in India, further data collection at local level needs to be undertaken. Second, this is a very short period of study (i.e., three months). For evaluating spatial and temporal patterns of the Covid pattern additional information is needed for a longer duration or more comprehensive chronological incidence.
6 Conclusion
In this study, GIS and spatial modelling have been used to analyze and show the spatial patterns of Covid through many epidemiological researches. The techniques Moran’s I and Getis-Ord G *i have been used spatial pattern and distribution pattern. The research findings indicated that the geographical distribution of Covid in India is heterogeneous, especially concentrated in the central and west of India. The findings of Covid hot spots in India (Maharashtra, Madhya Pradesh, Telangana and Rajasthan) with areal-based interpolation will help the provincial health officers to improve their remedial action and to establish potential strategies for better management of disease. Such spatial and temporal clusters can also try to empower and endorse highly efficient, locally adapted procedures for the highly spatially heterogeneous Covid disease. Similarly, our study indicates, by focusing where and when available public health resources should be focused, that spatial and temporal analyzes of population-based surveillance data for diseases will aid in manage viral diseases such as Covid.
References
Kamel Boulos, M. N., & Geraghty, E. M. (2020). Geographical tracking and mapping of coronavirus disease COVID-19/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic and associated events around the world: how 21st century GIS technologies are supporting the global fight against outbreaks and epidemics. International Journal of Health Geographics, 19, 8. https://doi.org/10.1186/s12942-020-00202-8.
Kumar, A. (2020). Modeling geographical spread of COVID-19 in India using network-based approach. medRxiv 2020.04.23.20076489. https://doi.org/10.1101/2020.04.23.20076489 Available at: https://www.medrxiv.org/content/10.1101/2020.04.23.20076489v1.full.pdf.
https://www.mygov.in/Covid-19. Accessed 23 May 2020.
Werneck, G. L. (2008). Georeferenced data in epidemiologic research. Ciência&SaúdeColetiva, 13(6), 1753–1766.
Subramanian, S. V., Karlsson, O., Zhang, W., & Kim, R. (2020). Geo-mapping of COVID-19 risk correlates across districts and parliamentary constituencies in India. Harvard Data Science Review. Retrieved from https://hdsr.mitpress.mit.edu/pub/zgoxm3ve.
Franch-Pardo, I., Napoletano, B. M., Rosete-Vergesa, F., & Billa, L. (2020). Spatial analysis and GIS in the study of COVID-19. A review. Science of the Total Environment, 739.
Robertson, C., & Nelson, T. A. (2014). An overview of spatial analysis of emerging infectious diseases. The Professional Geographer, 66(4), 579–588.
Tewara, M. A., Mbah-Fongkimeh, P. N., Dayimu, A., et al. (2018). Small-area spatial statistical analysis of malaria clusters and hotspots in Cameroon; 2000–2015. BMC Infectious Diseases, 18, 636. https://doi.org/10.1186/s12879-018-3534-6.
Xiong, Y., Guang, Y., Chen, F., & Zhu, F. (2020). Spatial statistics and influencing factors of the novel coronavirus pneumonia 2019 epidemic in Hubei Province, China. Research Square. https://doi.org/10.21203/rs.3.rs-16858/v2.
Goovaerts, P. (2005). Geostatistical analysis of disease data: estimation of cancer mortality risk from empirical frequencies using Poisson kriging. International Journal of Health Geographics, 4, 31. https://doi.org/10.1186/1476-072X-4-31.
Zarikas, V., Poulopoulos, S. G., Gareiou, Z., & Zervas, E. (2020). Clustering analysis of countries using the COVID-19 cases dataset. Data in Brief, 31, 105787.
Azevedo, L., Pereira, M. J., Ribeiro, M. C., & Soares, A. (2020). Geostatistical COVID-19 infection risk maps for Portugal. International Journal of Health Geographics, 19, 25. https://doi.org/10.1186/s12942-020-00221-5.
Moradi, M. M., Cronie, O., Rubak, E., et al. (2019). Resample-smoothing of Voronoi intensity estimators. Statistics and Computing, 29, 995–1010. https://doi.org/10.1007/s11222-018-09850-0.
Fisher, N. I., Lewis, T., & Embleton, B. J. J. (2016). Statistical analysis of spherical data, 1st edn. Cambridge: Cambridge University Press, 1987. Cambridge Books Online.
Yang, K., Li, W., Sun, L. P., Huang, Y. X., Zhang, J. F., Wu, F., et al. (2013). Spatio-temporal analysis to identify determinants of Oncomelaniahupensis infection with Schistosoma japonicum in Jiangsu province, China. Parasite & Vectors, 6(6), 138. https://doi.org/10.1186/1756-3305-6-138.PMID:23648203;PMCID:PMC3654978.
Naparus, M., & Kuntner, M. (2012). A GIS model predicting potential distributions of a lineage: A test case on hermit spiders (Nephilidae: Nephilengys). PLoS One, 7, e30047. https://doi.org/10.1371/journal.pone.0030047.
Samphutthanon, R., Tripathi, N., Ninsawat, S., & Duboz, R. (2013). Spatio-temporal distribution and hotspots of hand, foot and mouth disease (HFMD) in northern Thailand. International Journal of Environmental Research and Public Health, 11(12), 312–336.
Cliff, A. D., & Ord, J. K. (1981). Spatial processes: Models and applications. London: Pion.
Markoff, J., & Shapiro, G. (1973). The linkage of data describing overlapping geographical units. Historical Methods Newsletter, 7, 34–46.
Rosenshein, L. (2010). The local nature of a national epidemic: Childhood overweight and the accessibility of healthy food. M.S. dissertation, George Mason University, Department of Geography and GeoInformation Science, Fairfax, Virginia, USA.
Tobler, W. (1979). Smooth pycnophylactic interpolation for geographical regions. Journal of the American Statistical Association, 74, 519–530.
Lam, N. S. N. (1983). Spatial interpolation methods: A review. The American Cartographer, 10, 129–149.
SARS-CoV-2 (COVID-19) Testing. (2020). Status update. Indian Council of Medical Research. Retrieved 24 May 2020. Available at: https://www.icmr.gov.in/.
Tsai, P. J., Lin, M. L., Chu, C. M., & Perng, C. H. (2009). Spatial autocorrelation analysis of health care hotspots in Taiwan in 2006. BMC Public Health, 9, 464. https://doi.org/10.1186/1471-2458-9-464.
Desjardins, M. R., Hohl, A., & Delmelle, E. M. (2020). Rapid surveillance of COVID-19 in the United States using a prospective space-time scan statistic: Detecting and evaluating emerging clusters. Applied Geography, 102202.
Team CC-R. (2020). Geographic differences in COVID-19 cases, deaths, and incidence - United States, February 12–April 7, 2020. MMWR Morbidity and Mortality Weekly Report, 69(15), 465–471.
Silva, R. J., Silva, K., & Mattos, J. (2020). Análiseespacialsobre a dispersão da covid-19 no Estado da Bahia.
Santana Juárez, M. V. (2020). COVID-19 en México: Comportamientoespacio temporal y condicionantessocioespaciales, febrero y marzo de 2020 Posición, 3, 2683–8915.
Murugesan, B., Karuppannan, S., Mengistie, A. T., Ranganathan, M., & Gopalakrishnan, G. (2020). Distribution and trend analysis of COVID-19 in India: Geospatial approach. Journal of Geographical Studies, 4(1), 1–9.
Buchin, K., Buchin, M., van Kreveld, M., et al. (2012). Processing aggregated data: The location of clusters in health data. Geoinformatica, 16, 497–521. https://doi.org/10.1007/s10707-011-0143-6.
Arab-Mazar, Z., Sah, R., Rabaan, A. A., Dhama, K., & Rodriguez-Morales, A. J. (2020). Mapping the incidence of the COVID-19 hotspot in Iran – implications for travellers. Travel Medicine and Infectious Disease. https://doi.org/10.1016/j.tmaid.2020.101630.
Giuliani, D., Dickson, M. M., Espa, G., & Santi, F. (2020). Modelling and predicting the spatio-temporal spread of coronavirus disease 2019 (COVID-19) in Italy. SSRN. https://doi.org/10.2139/ssrn.3559569.
Roy, S., Bhunia, G. S., & Shit, P. K. (2020). Spatial prediction of COVID-19 epidemic using ARIMA techniques in India. Modeling Earth Systems and Environment, 1–7.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest between the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bhunia, G.S., Roy, S. & Shit, P.K. Spatio-temporal analysis of COVID-19 in India – a geostatistical approach. Spat. Inf. Res. 29, 661–672 (2021). https://doi.org/10.1007/s41324-020-00376-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41324-020-00376-0