Abstract
Introduction
Patients often take several different medications for multiple conditions concurrently. Therefore, when adverse drug events (ADEs) occur, it is necessary to consider the mechanisms responsible. Few approaches consider the mechanisms of ADEs, such as changes in physiological states. We proposed that the ontological framework for pharmacology and mechanism of action (pharmacodynamics) we developed could be used for this approach. However, the existing knowledge base contains little data on physiological chains (PCs).
Objective
We aimed to investigate a method for automatically generating missing PC from the viewpoint of anatomical structures. This study was conducted to determine dysuria-related adverse events more likely to occur during multidrug administration.
Methods
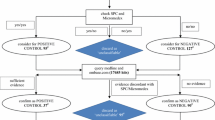
We adopted a systematic approach to determine drugs suspected to cause adverse events and incorporated existing data and data generated in our newly developed method into our ontological framework. The performance of automated data generation was evaluated using this newly developed system. Suspected drugs determined by the system were compared with those derived from adverse events databases.
Results
Of the 242 drugs involving suspected drug-induced urinary retention or dysuria, 26 suspected drugs were determined. Of these, five were drugs with side effects not listed in drug package inserts. The system derived potential mechanisms of action, PCs, and suspected drugs.
Conclusion
Our method is novel in that it generates PC data from anatomical structural properties and could serve as a knowledge base for determining suspected drugs by potential mechanisms of action.








Similar content being viewed by others

References
Arai H, Akishita M, Teramoto S, Mizukami K, Morimoto S, Toba K. Incidence of adverse drug reactions in geriatric units of university hospitals. Geriatr Gerontol Int. 2005;5:293–7.
Akishita M, Arai H, Arai H, Inamatsu T, Kuzuya M, Suzuki Y, et al. Survey on geriatricians’ experiences of adverse drug reactions caused by potentially inappropriate medications: Commission report of the Japan Geriatrics Society. Geriatr Gerontol Int. 2011;11:3–7.
Oosterhuis I, Zweers P, Rümke H, Muller-Hansma A, van Puijenbroek EP. A tailor-made approach for causality assessment for ADR reports on drugs and vaccines. Pharmacoepidemiol Drug Saf. 2018;38:1–7.
Campillos M, Kuhn M, Gavin A-C, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science (80-). 2008;321:263–6.
Garcia-Serna R, Mestres J. Anticipating drug side effects by comparative pharmacology. Expert Opin Drug Metab Toxicol. 2010;6:1253–63.
Chen B, Dong X, Jiao D, Wang H, Zhu Q, Ding Y, et al. Chem2Bio2RDF: a semantic framework for linking and data mining chemogenomic and systems chemical biology data. BMC Bioinform. 2010;11:255.
Sarntivijai S, Zhang S, Jagannathan DG, Zaman S, Burkhart KK, Omenn GS, et al. Linking MedDRA-coded clinical phenotypes to biological mechanisms by the ontology of adverse events : a pilot study on tyrosine kinase inhibitors. Drug Saf. 2016;39:697–707.
Berger SI, Iyengar R. Role of systems pharmacology in understanding drug adverse events. Wiley Interdiscip Rev Syst Biol Med. 2011;3:129–35.
Duran-Frigola M, Aloy P. Analysis of chemical and biological features yields mechanistic insights into drug side effects. Chem Biol. 2013;20:594–603.
Sider 2 [Internet]. 2015. http://sideeffects.embl.de/. Accessed 13 Mar 2015.
Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343.
Canadian Institutes of Health Research. DrugBank [Internet]. 2015. http://www.drugbank.ca/. Accessed 8 Dec 2018.
Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–6.
Stanford University. PharmGKB [Internet]. 2015. http://www.pharmgkb.org/. Accessed 13 Mar 2015.
McDonagh EM, Whirl-Carrillo M, Garten Y, Altman RB, Klein TE. From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomark. Med. 2011;5:795–806.
U.S. Department of Veterans Affair. National Drug File Reference Terminology (NDF-RT) [Internet]. http://www.va.gov/. Accessed 13 Mar 2015.
Lincoln MJ, Brown SH, Nguyen V, Cromwell T, Carter J, Erlbaum M, et al. US Department of veterans affairs enterprise reference terminology strategic overview. Stud Health Technol Inform. 2004;107:391–5.
Hayakawa M, Imai T, Ohe K. Investigation of descriptions in open drug information databases toward adverse events reasoning system. Jpn J Med Inform. 2013;33(Suppl.):898–901.
Zhichkin PE, Athey BD, Avigan MI, Abernethy DR. Needs for an expanded ontology-based classification of adverse drug reactions and related mechanisms. Clin. Pharmacol. Ther. 2012;91:963–5.
Gene Ontology Consortium. Gene Ontology [Internet]. 1999. http://geneontology.org/. Accessed 8 Dec 2018.
Foundational Model of Anatomy (FMA) [Internet]. 2019. http://si.washington.edu/projects/fma. Accessed 18 May 2019.
Imai T, Kou H, Zhou J, Kozaki K, Mizoguchi R, Ohe K. Japan Medical Ontology Development Project for Advanced Clinical. In: System Proceedings of 10th international HL7 interoperability conference 2009; 2009. p. 42–6.
Kozaki K, Yamagata Y, Mizoguchi R, Imai T, Ohe K. Disease compass—a navigation system for disease knowledge based on ontology and linked data techniques. J Biomed Semant. 2017;8:1–18.
Imai T, Ohe K, Shinohara E, Kajino M, Sakurai R, Kozaki K, et al. An ontological framework for representing topological information in human anatomy. In: Proc international conference on biomedical ontology BioCreative (ICBO-BioCreative 2016), Corvallis, USA, August 1–4, 2016 CEUR Work Proceedings, vol. 1747; 2016. p. 3–8. http://ceur-ws.org/Vol-1747/IT506_ICBO2016.pdf. Accessed 8 Dec 2018.
Mizoguchi R. YAMATO: Yet Another More Advanced Top-level Ontology [Internet]. 2016. http://www.ei.sanken.osaka-u.ac.jp/hozo/onto_library/upperOnto.htm. Accessed 18 May 2019.
Borgo S, Mizoguchi R. A first-order formalization of event, object, process and role in YAMATO. Front Artif Intell Appl. 2014;267:79–92.
Mizoguchi R. YAMATO: Yet Another More Advanced Top-level Ontology. In: Proceedings of sixth Australas ontology work Adelaide, Aust 7 December 2010; 2010. p. 1–16. https://pdfs.semanticscholar.org/7117/fa4b0fdaa737a90343ce8d4fcc890c57ea27.pdf. Accessed 8 Dec 2018.
Mizoguchi R, Toyoshima F. YAMATO : Yet Another More Advanced Top-level Ontology with Analysis of Five Examples of Change. In: Proceedings of the jointt ontological work 2017, Bozen-Bolzano, Italy, Sept 2017, vol. 2050. p. 21–23. http://ceur-ws.org/Vol-2050/FOUST_paper_4.pdf. Accessed 8 Dec 2018.
Kozaki K, Kitamura Y, Ikeda M, Mizoguchi R. Hozo: An environment for building/using ontologies based on a fundamental consideration of “role” and “relationship.” Knowl Eng Knowl Manag Ontol Semant Web. 2002;213–8. https://link.springer.com/chapter/10.1007/3-540-45810-7_21. Accessed 8 Dec 2018.
Mizoguchi R, Sunagawa E, Kozaki K, Kitamura Y. The model of roles within an ontology development tool: Hozo. Appl Ontol. 2007;2(2):159–79. https://content.iospress.com/articles/applied-ontology/ao038. Accessed 8 Dec 2018.
Kozaki K, Sunagawa E, Kitamura Y, Mizoguchi R. Role Representation model using OWL and SWRL. In: Proceedings of 2nd workshop on roles and relationships in object oriented programming, multiagent systems, and ontologies, Berlin, 30–31 July, p. 39–46, 2007. http://www.ei.sanken.osaka-u.ac.jp/pub/kozaki/Role07kozaki.pdf. Accessed 8 Dec 2018.
Yamagata Y, Kozaki K, Imai T, Ohe K, Mizoguchi R. An ontological modeling approach for abnormal states and its application in the medical domain. J Biomed Semant. 2014;5:23.
Imai T, Hayakawa M, Ohe K. Development of description framework of pharmacodynamics ontology and its application to possible drug-drug interaction reasoning. Stud Health Technol Inf. 2013;192:567–71.
Brunton LL, Chabner BA. Goodman and Gilman’s the pharmacological basis of therapeutics. New York: McGraw-Hill Prof. McGraw-Hill Professional; 2011. p. 178–80.
Tanaka C, Kato R. NEW pharmacology revised 6th edition. Nancodo: Tokyo; 2011. p. 234.
Fukuda K. Standard physiology revised 8th edition. Tokyo: Igaku-shoin; 2014.
The ICH MedDRA Management Committee Maintenance and Support Services Organization. MedDRA [Internet]. http://www.meddra.org/. Accessed 18 May 2019.
Brown EG. Using MedDRA implications for risk management. Drug Saf. 2004;27:591–602.
Japan Pharmaceuticals and Medical Devices Agency of. Safety Alerts and Recalls/Review Reports/Package Inserts, etc [Internet]. 2013. http://www.pmda.go.jp. Accessed 20 May 2013.
Zaman S, Sarntivijai S, Abernethy DR. Use of biomedical ontologies for integration of biological knowledge for learning and prediction of adverse drug reactions. Gene Regul Syst Biol. 2017;11:1177625017696075.
Therapeutic target database [Internet]. 2018. http://bidd.nus.edu.sg/group/cjttd/. Accessed 18 May 2019.
Yang H, Qin C, Li YH, Tao L, Zhou J, Yu CY, et al. Therapeutic target database update 2016: Enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016;44:D1069–74.
STITCH Consortium. STITCH [Internet]. 2016. http://stitch.embl.de/. Accessed 18 May 2019.
Kuhn M, Szklarczyk D, Pletscher-Frankild S, Blicher TH, Von Mering C, Jensen LJ, et al. STITCH 4: Integration of protein-chemical interactions with user data. Nucleic Acids Res. 2014;42:401–7.
European Molecular Biology Laboratory. ChEMBL [Internet]. 2018. https://www.ebi.ac.uk/chembl/. Accessed 18 May 2019.
Gaulton A, Hersey A, Nowotka ML, Patricia Bento A, Chambers J, Mendez D, et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45:D945–54.
EMBL-EBI. ChEBI [Internet]. 2018. http://www.ebi.ac.uk/chebi/. Accessed 18 May 2019.
Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–50.
Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–7.
Avillach P, Coloma PM, Gini R, Schuemie M, Mougin F, Dufour J-C, et al. Harmonization process for the identification of medical events in eight European healthcare databases: the experience from the EU-ADR project. J Am Med Inf Assoc. 2013;20:184–92.
Xu R, Wang Q. Automatic signal extraction, prioritizing and filtering approaches in detecting post-marketing cardiovascular events associated with targeted cancer drugs from the FDA Adverse Event Reporting System (FAERS). J Biomed Inform. 2014;47:171–7.
Liu Z, Fang H, Reagan K, Xu X, Mendrick DL, Slikker W, et al. In silico drug repositioning: what we need to know. Drug Discov Today. 2013;18:110–5.
Jung K, LePendu P, Chen WS, Iyer SV, Readhead B, Dudley JT, et al. Automated detection of off-label drug use. PLoS One. 2014;9:e89324.
Yildirim MA, Goh K-I, Cusick ME, Barabási A-L, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–26.
Hisaka A, Ohno Y, Yamamoto T, Suzuki H. Prediction of pharmacokinetic drug-drug interaction caused by changes in cytochrome P450 activity using in vivo information. Pharmacol Ther. 2010;125:230–48.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Research involving human participants and/or animals
This manuscript does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
Masayo Hayakawa, Takeshi Imai, Yoshimasa Kawazoe, Kouji Kozaki, and Kazuhiko Ohe have no conflicts of interest that are directly relevant to the content of this study.
Funding
This work was partially supported by Health, Labour and Welfare Sciences Research Grants number H28-IRYO-SHITEI-020, Japan, and JSPS KAKENHI Grant number JP26460857.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hayakawa, M., Imai, T., Kawazoe, Y. et al. Auto-Generated Physiological Chain Data for an Ontological Framework for Pharmacology and Mechanism of Action to Determine Suspected Drugs in Cases of Dysuria. Drug Saf 42, 1055–1069 (2019). https://doi.org/10.1007/s40264-019-00833-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40264-019-00833-2