Abstract
Key message
A moderate genetic diversity, the absence of a significant genetic differentiation between wild and cultivated stands and a highly admixed genetic structure of sweet chestnut with two main clusters were observed in France using two different data sets with 10 and 18 microsatellites.
Context
Renewed interest in European chestnut in France is focused on finding locally adapted populations partially resistant to ink disease and identifying local landraces.
Aims
We genotyped trees to assess (i) the genetic diversity of wild and cultivated chestnut across most of its range in France, (ii) their genetic structure, notably in relation with the sampled regions, and (iii) to a lesser extent the relations between French chestnuts with 10 cultivated chestnuts from the Northwest of Spain that were previously classified in the Iberian or Italian groups.
Methods
A total of 693 trees in 16 sampling regions in France were genotyped at 24 SSRs and 1401 trees in 17 sampling regions at 13 SSRs.
Results
Genetic diversity was moderate in most sampling regions, with redundancy between them. No significant differentiation was found between wild and cultivated chestnut. A genetic structure analysis with no a priori information found a low yet significant structure and identified two clusters at 18 SSRs. One cluster gathers trees from south-east France and Corsica (RPP1), and another cluster gathers trees from all other sampled regions (RPP2). A substructure was detected at 10 SSRs suggesting differentiation within both RPP1 and RPP2. RPP1 was split between south-east France and Corsica. RPP2 was split between northwest France, Aveyron, Pyrenees, and a last cluster gathering individuals from several other regions.
Conclusion
The genetic structure within and between our sampling regions is likely the result of natural events (recolonization after the last glaciation) and human activities (migration and exchanges). These advances on our knowledge of chestnut genetic diversity and structure will benefit conservation and help our local partners’ valorization efforts.
Similar content being viewed by others
1 Introduction
Sweet chestnut (Castanea sativa Mill.) is an endemic, multi-purpose tree species cultivated for its wood and nuts. It is the third broad-leaved tree species in France in forest area (750,000 ha) and in 2016 accounted for 5% of land used for fruit production (FranceAgriMer 2017). With an annual production of 7000–9000 t in the last 10 years, France is the fifth European producer (FAO 2018). Sweet chestnut has been intensively cultivated in coppices and orchards for centuries in France. However, since the beginning of the eighteenth century, it has suffered from abandonment, leading to a sharp decrease in production (Pitte 1986; Sauvezon et al. 2000). Many landraces and associated knowledge were lost. In the early twentieth-century, the Laffite tree nursery in Basque Country started to breed hybrid chestnut followed in the 1960s by the French National Institute for Agricultural Research (INRA) (Pereira-Lorenzo et al., 2016). These breeding programs aimed at developing interspecific hybrids resistant to ink disease caused by a Phytophthora oomycete, by crossing two Asiatic tolerant species, Castanea crenata and Castanea mollissima, with local landraces from regions with an oceanic climate. These hybrids are now mainly used for fruit production and as rootstock (particularly Marigoule and Bouche de Bétizac varieties and more recently BelleFer). However, they are not adapted to continental and Mediterranean conditions (Martín et al. 2017; Míguez-Soto et al. 2019). Their fruit quality has also been criticized by some growers and by chestnut lovers, particularly in comparison with landraces. Action was thus taken by these actors, involving survey of old chestnut trees, phenotypic observations, and the establishment of conservatory orchards.
Strong geographical structure was reported in wild populations in Italy, Spain, Greece, and Turkey (Mattioni et al. 2013). A study of wild chestnut in Spain, Italy, and Greece (Fernández-Cruz and Fernández-López 2016) found two main gene pools in Europe, and another study of wild, natural, or naturalized populations across Europe (Mattioni et al. 2017) found three. These findings agree with evidence of spontaneous establishment originating from the Last Glacial Maximum refugia in the North of the Iberian, Italian, and Balkan peninsulas and in northern Anatolia (Krebs et al. 2004, 2019; Roces-Díaz et al. 2018). In southern France, there is possible evidence for chestnut refugia in palaeobotanical data (Krebs et al. 2019). The preferred hypothesis is therefore that most pre-cultivation Castanea in France are the result of the spontaneous spread of the species from neighboring southern European refugia, i.e., in Spain and Italy. However, the most recent genetic analyses conducted exclusively on French populations were published in the 1990s on wild chestnut and at a regional scale (Frascaria et al. 1991, 1992; Frascaria and Lefranc 1992), and the results obtained in the CASCADE project (Eriksson et al. 2005) have not yet been published (T. Barreneche pers. com.). Mattioni et al. (2008) compared naturalized, coppice, and orchard populations in Italy, Greece, Spain, the UK, and France and showed differences in within-population genetic parameters between fruit orchards and other types of chestnut management. This result implies that long-term management techniques can influence the genetic makeup of the populations. Differences between and within countries have also been reported (Pereira-Lorenzo et al. 2016). For these reasons, specifically French, finer-scale sampling of both wild (forest) and cultivated chestnut trees (orchards and alignments) was needed to help distinguish between natural and anthropogenic evolutionary factors.
In terms of sampling, many authors have genotyped tree collections ex situ, i.e., in conservatories (Martín et al. 2010a), and in situ (Pereira-Lorenzo and Fernandez-Lopez 1997; Gobbin et al. 2007; Martín et al. 2010b; Pereira-Lorenzo et al. 2010, 2019; Beccaro et al. 2012; Mellano et al. 2012, 2018; Beghè et al. 2013; Quintana et al. 2014; Fernández-López and Fernández-Cruz 2015). In this study, we used both in- and ex situ sources to assess the known and currently used genetic diversity of sweet chestnut. As a result, we often sampled several individuals belonging to the same landrace. Hereafter, we use the term “landrace” as defined by Villa et al. (2005) rather than “variety” or “cultivar,” as it better covers the variety of sampling situations we encountered in the field. However, we do use the term “cultivar” when known cultivars were encountered.
The main aims of this work were to assess (i) the genetic diversity of wild and cultivated chestnut in most of its range in France, (ii) their genetic structure, notably in relation with the sampling regions, and (iii) to a lesser extent the relations between French chestnuts with 10 cultivated chestnuts from the northwest of Spain that were previously classified in the Iberian or Italian groups. For this purpose, we sampled natural chestnut populations, ancient grafted chestnut identified in situ by local partners and ex situ local landraces in conservatories in the main nut-producing regions, and in most of the distribution of natural chestnut forests in France. We used microsatellite markers from the EU chestnut database to genotype all sampled trees at 13 SSRs and a subset at 24 SSRs (Pereira-Lorenzo et al. 2017). By also including Iberian samples cited in the Pereira-Lorenzo et al. publication, we also provide some evidence for the origin of the trees we sampled.
Fixation of genotypes by grafting from spontaneous chestnut, or “instant domestication” as defined by (Harris et al. 2002), is reported in the literature (Aumeeruddy-Thomas et al. 2012) and was recently documented in Italy and Spain (Pereira-Lorenzo et al. 2019). As a working hypothesis, this suggested a possible lack of genetic structure between wild and cultivated chestnut. It is common knowledge that grafts and nuts travel by means of markets, historically via occupational travelers such as glass blowers (Pitte 1986) and now via local and internet-mediated exchange fairs. However, the extent and impact of this phenomenon on the genetic structure of cultivated chestnut were previously unknown in France. We hypothesized that it is sufficiently frequent to have a significant impact, leading to a low genetic structure of cultivated chestnut in France. As reported in Pereira-Lorenzo et al. (2019), we also expected to find a high overall genetic diversity but without marked differences between the wild and cultivated sets. In addition, we expected in situ local landraces to be multiclonal due to repeated grafting over the centuries and the accumulation of mutations or the use of seedlings from these landraces.
2 Materials and methods
Terminology: we avoid the use of “population” and instead use “sampling region” to describe a geographically or socially meaningful region where a non-profit association has prospected and conserved chestnut or a group of sampling sites located close by. We use “genetic cluster” to denote a cluster of genotyped trees resulting from the analysis of their genetic structure. “Chestnut type” is used as a category with two levels, “forest” and “cultivated.”
2.1 Sampling scheme
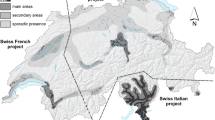
To characterize and understand the genetic diversity and population structure of the European chestnut (C. sativa Mill.) in France, we genotyped 693 trees in 16 sampling regions in both forest and cultivated areas. Table 1 lists sampling details, and Fig. 1 shows the location of the sampling regions (GPS of sampled trees are available upon request). We added the genotyping of 12 more Spanish trees to allow the future insertion of the French genotypes into the European database (Pereira-Lorenzo et al. 2017).
Map of sampling regions. The distribution of chestnut forest areas where chestnut accounts for at least 75% of the leaf cover, which represents about 50% of the total chestnut-comprising forest area in France (IGN 2007) (in green) and the distribution of cultivated chestnut and orchards (IGN 2016) (in orange). Each dot represents a sampling region where chestnut forest or orchard is present (this qualitative information does not reflect the relative areas or number of trees)
2.2 Geographical sampling
In forest stands, trees were chosen randomly, located several dozen meters apart in the middle of forest patches. Their exact locations were recorded by GPS. In Brittany, Auvergne-Rhône-Alpes, Occitanie, Provence-Alpes-Côtes d’Azur (PACA), and Corsica, mature leaves were sampled and immediately enclosed in plastic bags with silica gel. In Gironde, dormant buds were sampled from trees close to the laboratory to facilitate frequent resampling when assessing the accuracy of genotyping protocols. In Corsica, nuts and dried leaves were also sampled in the field, whereas cultivated chestnut was provided as DNA extract. Whenever we sampled offspring as groups of half sib fruits, we also sampled leaves from their mothers. Nuts harvested in the Finistère, Corsica, Basque Country, and Aveyron forest sampling regions were germinated and sown in the greenhouse.
2.3 Expert-based sampling
Field surveys of cultivated chestnut were conducted in 2016–2017 in collaboration with producer and amateur organizations. In 2016, we focused our sampling effort on the landraces they knew and were interested in. In 2017, we expanded sampling to most known landraces and grafted trees, supplemented by random sampling in a few chestnut orchards. Associative conservatories were also sampled. We sampled several chestnut trees that had the same name to test the genetic diversity of landraces. When attributing sampled trees to a given landrace, when known, we followed the field expert’s determination.
2.4 SSR genotyping
A total of 693 trees were genotyped at 24 SSRs and 1401 trees at 13 SSRs. Total genomic DNA was extracted from fresh leaves, silica-dried leaves, or dormant buds using the DNeasy 96 Plant kit (Qiagen, Hilden, Allemagne). Twenty-four SSR markers previously selected to study chestnut genetic diversity were used for this study (Buck et al. 2003; Gobbin et al. 2007; Kampfer et al. 1998; Marinoni et al. 2003; Steinkellner et al. 1997) based on the protocol of Pereira-Lorenzo et al. (2017). We amplified these 24 SSRs into 5 multiplex and 2 singleplex PCRs using one of the FAM, NED, PET, and VIC fluorophore-labeled primers (PE Applied Biosystems, Warrington, UK) modified following Pereira-Lorenzo et al. (2017, 2019). The PCR final reaction volume was 15 μl (7.5 μl of QIAGEN Multiplex Master Mix, 0.075 to 0.3 μM of each primer, 4 to 4.9 μl RNase Free Water, and 2 μl of ADN at 5–10 ng/μl). The amplification conditions were 95 °C for 5 min, followed by 30 cycles at 95 °C for 30 s, annealing at a specific temperature depending on the multiplex set, for 1.5 min and 1 min at 72 °C, and final extension at 60 °C for 30 min. Negative controls were included in all PCR reactions to enable detection of cross-contamination of the samples. Amplifications at 13 SSRs corresponded to sets 1, 2, and 3. Amplifications at 24 SSRs corresponded to all sets. The descriptions of all sets are displayed in Table 2. Amplification products were diluted with water, and 2 μl of the diluted amplification product was added to 0.12 μl of 600LIZ size standard (Applied Biosystems, Foster City, USA) and 9.88 μl of formamide.
Genotyping was performed partly on an ABI 310 capillary sequencer (Applied Biosystems, Foster City, CA, USA) at the Xylobiotech FCBA facility of Cestas-Pierroton with further work on an ABI 3500 XL capillary sequencer (Applied Biosystems, Foster City, CA, USA) at the CIRAD GenSeqUM platform in Montpellier, France. Allele sizes were read independently by two investigators using GENEMAPPER 4.1 and 5.0, respectively (Applied Biosystem, Foster City, USA). The output files in the fsa format were made compatible for GENEMAPPER 4.1 using a Python script from the Montpellier platform.
2.5 Data analysis
2.5.1 Data filtering: detection of interspecific hybrids, clonal groups, and null alleles
All individuals with more than 20% of missing alleles were removed along with individuals showing Asiatic alleles. Asiatic genotypes present initially in our data, and Asiatic alleles identified in a previous study (Pereira-Lorenzo et al. 2010) were used to detect exotic germplasm in the European populations.
The presence of uninformative loci was tested with the informloci function in the R/poppr package version 2.8.3 (Kamvar et al. 2015; Kamvar et al. 2014) in both data sets. The percentages of missing data were obtained using the info_table function in R/poppr. The frequency of null alleles per locus was calculated with the R/PopGenReport package version 3.0.4 (Adamack and Gruber 2014) based on Brookfield formula (Brookfield 1996). Following Lassois et al. (2016), we discarded loci with more than 10% of null alleles. After removing loci, the genotype curve function implemented in the R/poppr. was applied to both data sets to determine the minimum number of loci necessary to discriminate between individuals. Repeated genotypes were searched within each sampling region to identify multi-locus genotypes (MLGs) for each data set, using the clonecorrect function in R/poppr. When detected, these repeated MLGs were removed. Note that some MLGs may be repeated between sampling regions, but those were kept for the rest of the analyses as they provide information on population structure.
2.5.2 Genetic diversity
The observed number of alleles (Na), expected heterozygosity (He), and observed heterozygosity (Ho) were calculated at each locus across sampling regions, per sampling region, and per genetic cluster, using the summary function in the R/adegenet package 2.1.1 (Jombart 2008). The Fst and corrected Fst (Fstp) and Fis and Dest per locus (Jost 2008; Nei 1987) were calculated using the basic.stats function in the R/hierfstat package version 0.04-22 (Goudet 2005). The poppr function in R/poppr was used to report other basic statistics per sampling region including the Shannon-Weiner diversity index (H), the index of association (Ia), and the standardized index of association (rbarD) and the effective number of alleles (Ne) with Ne = 1/(1-He) (Kimura and Crow, 1964; Jost, 2008). The significance of Ia and rbarD was tested with 1000 permutations, shuffling the genotypes at each locus while maintaining the heterozygosity and allelic structures. The Fis per sampling regions and per genetic cluster was calculated using the basic.stats function in the R/hierfstat package version 0.04-22 (Goudet 2005). Bootstrapping of Fis per sampling regions and per genetic cluster was calculated using the boot.ppfis function in the R/hierfstat package version 0.04-22 (Goudet 2005). Deviation from the Hardy-Weinberg equilibrium (HWE) was tested on both loci and populations with 1000 permutations using the hw.test function in the R/pegas package version 0.11 (Paradis 2010). The Chi2 statistic was calculated over the entire data set, and two p values were computed, one analytical and one derived from 1000 Monte Carlo permutations. The significance threshold was set at 0.05.
2.5.3 Population structure
For each data set, three methods were compared to find genetic clusters, by varying the number of clusters, K, between 1 and 15: STRUCTURE (Pritchard et al., 2000; Falush et al., 2003), discriminant analysis of principal components (DAPC: Jombart et al., 2010), and sparse non-negative matrix factorization (SNMF: Frichot et al., 2014). For the first method, the STRUCTURE software v2.3.4 was launched thirty times for each K value using the admixture model with unlinked loci and correlated frequencies (other parameters: usepopinfo = 0, popflag = 0, burn-in = 200,000, MCMC iterations = 200,000). The number of clusters was determined using the DeltaK metric from structureHarvester v0.6.94 (Evanno et al., 2005). At the best K, the assignment probabilities of each individual to each cluster (Q matrix) were averaged over the 30 runs using a suitable permutation to handle label switching. The permutations were obtained with the CLUMPP software v1.1.2 using the LargeKGreedy algorithm with 2000 repeats (Jakobsson and Rosenberg, 2007). For the second method, SSR genotypes were first transformed by a principal component analysis (PCA), followed by the K-means algorithm applied to the principal components using the find.clusters function in R/adegenet v2.1. The number of clusters was determined using the BIC. The DAPC was then performed at the best K with the number of principal components to keep chosen by cross-validation using the xvalDapc function in R/adegenet with 30 repetitions and a maximum of 300 PCs. For the third method, the snmf function of R/LEA v2.0 was launched with 5 repetitions. The number of clusters was determined using the cross-entropy criterion. For all methods, the clusteredness metric was computed to assess the extent to which a randomly chosen individual is inferred to have ancestry in only one cluster (Rosenberg et al., 2005). The threshold above which individuals were considered as “strongly assigned” was set to 0.8 (qI ≥ 80%).
Hierarchical analysis of molecular variance (AMOVA, Excoffier and Smouse 1992) as implemented in the poppr.amova function in R/poppr was performed using all loci with less than 5% missing data on the preset hierarchy of chestnut types and sampling regions and on genetic clusters. Fis, pairwise Fst, and hierarchical F-statistics were calculated, and 95% confidence intervals were obtained by bootstrapping with 1000 samples over loci using the boot.ppfis, boot.ppfst, and boot.vc functions. Differences between hierarchy levels were tested by randomization with the function randtest in the R/ade4 package version 1.7-13 (Excoffier and Smouse 1992; Chessel et al. 2004). Some components of covariance could have slightly negative estimates due to the absence of significant genetic structure at the corresponding hierarchical level (FAQ List for Arlequin 2000).
2.5.4 Reproducibility
To facilitate method reproducibility (Goodman et al. 2016), most of our analyses were performed in R (R Core Team 2019); the scripts are available on the server data.inrae.fr (Bouffartigue et al., 2020).
3 Results
3.1 Data filtering: detection of interspecific hybrids, clonal groups, and null alleles
For the 24 SSRs data set, 13 genotyped trees with more than 20% of missing alleles were filtered out (respectively, 32 trees at 13 SSRs). Three trees genotyped twice were filtered out at 24 SSRs (respectively, 18 trees at 13 SSRs). Over all our data sets, Asiatic alleles were scored as following: CsCAT41A-196, EMCs15-76, EMCs2-152, QpZAG36-205, QrZAG4-118, QrZAG96-154, EMCs14-135, CsCAT14-136, CsCAT6-138, CsCAT1-188, CsCAT17-135, CIO-160, OCI-160, OCI-167, OCI-169 and OAL-307, CsCAT1-188, CsCAT6-138, CsCAT17-135, CIO-160, CIO-167; CIO-169 and OAL-307. For the 24 SSR data set, 26 interspecific trees were filtered out, 11 were known beforehand and 15 were detected out of specific alleles known from Asiatic species (respectively, 127 trees at 13 SSRs: 18 were known beforehand and 119 were detected out of known Asiatic alleles). Most of the detected interspecific trees were from ForBasque. Nine Spanish trees were filtered out at 24 SSRs (respectively, 10 trees at 13 SSRs). A total of 51 trees were discarded at 24 SSRs (respectively, 187 trees at 13 SSR). For the 24 SSRs data set, 642 trees genotyped (respectively, 1214 at 13 SSRs) remained for further analysis (Table 1).
CsCAT41 is known to amplify two sites: CsCAT41A and CsCAT41B. CsCAT41A was less polymorphic, with only 2 alleles detected in our analyses out of 3 already known (Pereira-Lorenzo et al., 2010): CsCAT41A-196 which matches with CsCAT41A-200 in Pereira-Lorenzo et al. (2010) and CsCAT41A-199 which matches with CsCAT41A-202 in Pereira-Lorenzo et al. (2010). CsCAT41A-188 in Pereira-Lorenzo et al. (2010) did not came out in our data set. CsCAT41A was removed from the subsequent analyzes. CsCAT41B was more polymorphic, with 11 alleles (210–233) in the data set with 24 SSRs and 13 alleles (210–237) in the data set with 13 SSRs.
After filtering for null alleles, EMCs38, CIO, and EMCs25 were discarded from the data set at 24 SSRs (respectively, EMCs38 at 13 SSRs) (ESM_1). EMCs14, EMCs2, and EMCs25 had more than 5% of missing alleles and were discarded from the data set at 24 SSRs (respectively, QrZAG4 at 13 SSRs). The non-phylogenetically informative loci EMCs14 was discarded from both data sets (minor allele frequency MAF < 0.01).
The resulting data sets had 18 SSRs, hereafter called 18All, and 10 SSRs, hereafter called 10All. Repeated multi-locus genotypes (MLGs) were then detected in each sampling region, as they could be the result of both practices (grafting) and sampling choices. They had to be removed to avoid the artefactual detection of genetic structure resulting from the sampling strategy. The resulting data sets (Table 1) are called 10Unik (1050 trees) and 18Unik (520 trees). Four MLGs among sampling regions were detected at 24 SSRs (respectively, 9 at 13 SSRs) but were not discarded as they provide information on populations’ structure. In both data sets, the discriminating power of the polymorphic markers to differentiate between genotypes was sufficient to discriminate all individuals irrespective of the number of loci and individuals (ESM_2).
3.2 Description of SSR diversity per sampling region
The 18Unik data set analyzed in this study varied greatly in allele diversity (ESM_3A). The 18Unik data set (respectively, 10Unik; see ESM_3B) had a total of 179 alleles (respectively, 112) with an average of 9.9 alleles per locus (respectively, 11.2). This ranged from 2 for QrZAG4 to 31 for CsCAT3 (respectively, 3 for EMCs2 to 33 for CsCAT3). In terms of expected heterozygosity (He), QrZAG4 showed the lowest diversity with 0.17 in 18Unik (respectively, EMCs2 with 0.66 in 10Unik) and CsCAT6 the highest diversity with 0.86 in 18Unik (respectively, CsCAT3 with 0.85 in 10Unik). The within-population inbreeding coefficient (Fis) ranged from − 0.427 to 0.153 in 18Unik (respectively, − 0.431 to 0.169 in 10Unik), with a mean of − 0.102 in 18Unik (respectively, − 0.031 in 10Unik). In 18Unik, only QpZAG110, QrZAG4, and RIC were in the HWE (ESM_4A). Across all sampling regions, in 10Unik, none of the SSRs were in HWE at both probabilities (ESM_4C). When tested per sampling region, only ForGard and ForBasque were in the HWE in both data sets. ForAveyron was in the HWE only in 10Unik. Moreover, in both data sets, HWE was rejected for all SSR loci in at least one sampling region except OCI in 18Unik
3.3 Redundant diversity among sampling regions and no differentiation between chestnut type
Genetic diversity indices calculated for each sampling region genotyped at 18 SSRs without MLGs are listed in Table 3 (results at 18 SSRs are presented in ESM_5). Note that the aim of sampling ForGironde was not to be representative of the region but to facilitate resampling. Moreover, in the 18Unik data set, ForBasque had a single individual. Therefore, diversity and differentiation are discussed excluding ForGironde and ForBasque in the 18Unik data set and excluding ForGironde in the 10Unik data set. The highest effective number of alleles per sampling region was found in the Finistère forest sampling regions in 18Unik (ForFinistere, northwestern France), and the lowest was found in the cultivated sampling region in Var (CultVar, south-east of France). The mean observed heterozygosity was 0.671, and the mean expected heterozygosity was 0.605. The sampling regions with the lowest (respectively highest) observed heterozygosity were ForAveyron, the forest sampling region in Aveyron (respectively, CultVar). The sampling regions with the lowest (respectively highest) expected heterozygosity were CultVar (respectively, the forest sampling regions in Finistère, ForFinistere). Excluding ForGironde, positive and significant inbreeding (Fis) was found in the cultivated sampling region in Ariège (CultAriege) and in the forest sampling region in Ardèche (ForArdech). The highest Ia and rbarD were found in CultVar, and the lowest were found in ForFinistere. The results of AMOVA (Table 4 and ESM_6A) revealed no substantial difference in structure in chestnut type between forest stands and cultivated orchards: the variance component did not significantly differ from zero. Moreover, although the Fct had a confidence intervals excluding zero, it is very close to zero. Instead, more than 80% of the variance was found within each sampling region. At a threshold of 0.001, we rejected the null hypothesis of panmixia, both among sampling regions within chestnut types and within sampling regions. Among sampling regions within chestnut types, the Phi test statistic of the AMOVA indicated greater variance than expected under the null hypothesis. This suggested an underlying structure at this hierarchical level that was confirmed by a positive bootstrap-derived confidence interval for Fst (0.088–0.11). Within sampling regions, the Phi test statistic indicated lower variance than expected under the null hypothesis. This suggested some inbreeding at this hierarchical level confirmed by a negative bootstrap-derived confidence interval for Fis, although very close to zero. Note that in the 10Unik data set, a bootstrap-derived confidence interval for Fis included zero (ESM_6A).
3.4 Two main genetic clusters with a strong admixture among sampling regions
In addition to analyzing genetic diversity per sampling region, we also evaluated the overall genetic structure to detect genetic clusters, if any, and to assess their congruence with respect to the sampling regions.
The number of genetic clusters was determined using three methods, each with its own criterion: DeltaK for STRUCTURE, BIC for DAPC, and cross-entropy for SNMF. For all data sets, BIC and cross-entropy started by decreasing sharply (ESM_7), demonstrating the presence of genetic structure. However, the signal was not so clear, making the choice of the number of genetic clusters rather difficult. By contrast, the signal of DeltaK was clear for all data sets. Therefore, we chose to display in the main and supplementary material the results of STRUCTURE rather than the other methods.
On the 18Unik data set (respectively, 10Unik), the most likely number of clusters, according the DeltaK criterion, gave the highest value for K = 2 (respectively, K = 6). We chose to display the results at K = 2 and K = 6 for the 10Unik (ESM_7B) and 10Unik with Spanish sample data sets (ESM_7D) in order to allow comparison with the 18Unik (ESM_7A) and 18Unik with Spanish sample data set (ESM_7C).
Reconstructed panmictic populations (RPPs) were numbered according to the clustering of the 18Unik data set at K = 2 (RPP1 and RPP2), and the clustering of the 10Unik data set was named in reference to the K = 2 clustering (RPP1a, RPP1b, RPP2a, RPP2b, RPP2c, and RPP2d). In tables and electronic supplementary material (ESM), the numbers of clusters were kept together with the RPPs in order to help the reader to follow the results from both the manuscript and the files hosted on INRAE server.
On the 18Unik (Fig. 2), (respectively, 10Unik; see ESM_8B) data set, for K = 2, 85% (respectively, 85%) of the individuals from Corsica, Var, and Ardèche were grouped in reconstructed panmictic population 1 (RPP1), and 68% (respectively, 62%) of individuals from all sampling regions except Var (respectively, except the forest region in Var) were grouped in RPP2. Cultivated sampling region from Ardèche (CultArdech) and forest sampling region from Hérault (ForHerault) had more than 50% of admixed individuals between RPP1 and RPP2 (Fig. 2b). On the 10Unik data set, for K = 6, 95% of the individuals from Var and 20% of individuals from Ardèche were in RPP1a, 94% of the individuals from Corsica were grouped in RPP1b, 70% of individuals from Limousin and Finistère (west of France) were grouped in RPP2a, 71% of individuals from Aveyron and Cantal (south-west of Massif Central) were grouped in RPP2b, 68% of individuals from Ardèche were grouped in RPP2c, and 80% of individual from Ariège, Hautes-Pyrénées, and Basque Country (French Pyrennées) and 69% of individuals from Hérault were grouped in RPP2d. Cultivated sampling region from Ardèche (CultArdech), Ariège (CultAriege), Aveyron (CultAveyron), and Corsica (CultCorsica) and forest sampling region from Cantal (ForCantal), Finistère (ForFinistere), and Hérault (ForHerault) had more than 50% of admixed individuals between RPPs (ESM_8B for K = 6).
a Classification of 520 European chestnut genotypes, in reconstructed panmictic populations (RPPs) when K = 2, based on the 18Unik data set. In orange, genotypes from south-east of France and Corsica (RPP1). In green, genotypes from other regions (RPP2). b Table of assignment of sampling regions in RPPs when K = 2, based on the 18Unik data set. c Tables of pairwise Fst between RPPs using all individuals and strongly assigned individuals (ql ≥ 80%), when K = 2, based on the 18Unik data set
The overall admixed genetic structure in our sample was confirmed by the relatively low pairwise Fst calculated between clusters, as can be seen in Fig. 2 and ESM_8B: 0.09 between RPP1 and RPP2 when ql ≥ 80% on the 18Unik (respectively, 0.088 on the 10Unik) data set. When K = 6 and ql ≥ 80% on the 10Unik data set, the highest Fst values were found between Var and Ardèche (RPP1a) and Aveyron (RPP2b), West of France (RPP2a), and Pyrénées (RPP2d). The lowest Fst values were found between RPP1b, RPP2b, and RPP2d. Taking into account the confidence interval, the genetic differentiation between Var and Ardèche (RPP1a) and Corsica (RPP2b) is of the same magnitude than the genetic differentiation between Var and Ardèche (RPP1a) and Aveyron (RPP2b), West of France (RPP2a), and Pyrénées (RPP2d).On the 18Unik (respectively, 10Unik) data set, eighty-five individuals out of 520, i.e., 16% (respectively, 564 out of 1050, i.e., 54%) had a ql < 80%. A hierarchical AMOVA performed on the strongly assigned individuals (ql ≥80%) of the 18Unik data set (respectively, 10Unik) corroborated this finding (Table 5 and ESM_6C): 86.6% of the variance (respectively, 81.9%) was found among samples within clusters. For the 18Unik (respectively, 10Unik) data set, the variance component between clusters was 13.4% (respectively, 18.1%) and the Fst was of 0.09 (respectively, 0.125), indicating a relatively low but significant genetic structure (Table 5 and ESM_6C).
When the inbreeding coefficient was calculated per cluster on strongly assigned individuals (Table 6 and ESM_9), the 95% confidence interval of both clusters included zero. The mean observed heterozygosity was 0.673 for 18Unik (respectively, 0.657 for 10Unik), and the mean expected heterozygosity was 0.681 (respectively, 0.654 for 10Unik).
In terms of assignment probabilities of individuals to clusters, the “clusteredness” statistic was markedly higher for STRUCTURE and DAPC than for SNMF, for both 18Unik at K = 2 (0.809 and 0.909 versus 0.642) and 10Unik at K = 6 (0.732 and 0.922 versus 0.512).
4 Discussion
4.1 Sampling and genotyping
This work is the first comprehensive survey of genetic diversity and structure of C. sativa Mill. in France. In the 1990’s, Frascaria and colleagues sampled chestnut trees at four locations in France, and they used three isozymes loci. In terms of geographical sampling, we sampled trees at more locations partially including theirs and extending it to new locations. As a result, our sampling covers all the main areas of cultivated and forest chestnuts in France. Moreover, our use of DNA-based markers (SSR) presents several advantages over protein-based ones (used by Frascaria et al., 1992) as such markers provide a higher polymorphism per loci, hence a better discriminative ability (Gibson, 2014).
The markers we used were selected after an extensive review of the literature (by us for the 13 SSR and independently by Pereira-Lorenzo et al. (2017), and allele scoring was the subject of a recent optimization by Pereira-Lorenzo et al. (2017). Nevertheless, we faced the usual difficulties and drawbacks of microsatellites, i.e., errors and uncertainties in allele calling, difficulty in data comparison and transferability across labs and collaborators over time, and the huge amount of time needed to perform the analysis, as emphasized in previous studies (reviewed by Guichoux et al. 2011).
We consequently set up a small project to define nuclear SNPs, at least to check clear duplicates (in the case of good quality genotyping results) and putative duplicate (in the case of low-quality results) among samples from variety repositories. In a few months, we re-genotyped about 500 samples with up to 160 SNPs and confirmed all suspected duplicates. A detailed description of this work will be the subject of a separate article.
4.2 Null allele frequencies and genetic diversity per sampling region
Some SSRs were known to often have a high null allele frequency, such as EMCs25 (Lusini et al. 2013) and CsCAT14, CsCAT2, CsCAT41B, QrZAG4, and CIO (Pereira-Lorenzo et al. 2017). In our data sets, EMCs38 had between 14 and 19 alleles, whereas CIO had 6 and EMCs25 had 7. These two loci likely had a high null allele frequency because of an excess of homozygotes.
Jost (2008) convincingly argued that the effective number of alleles, Ne, is a more relevant metric of diversity than heterozygosity. The range of this diversity metric in our sampling regions (2.53–3.31) is markedly higher than what Mattioni et al. (2008) obtained on their samples from France (1.33) as well as their samples from Spain (1.30) and Italy (1.35). However, it is similar or even lower than the ones from Mattioni et al. (2013) for Spain (3.12–3.41) and Italy (2.83–4.93) and from Mattioni et al. (2017) for France (3.46–3.82), Spain (3.06–4.31), and Italy (3.24–5.57). In comparison, Skender et al. (2017) obtained a Ne range of 2.15–2.75 in Bosnia-Herzegovina and Croatia. Such comparisons suggest that our sampling regions in France have an intermediate level of diversity, likely similar (resp. lower) to those sampled by colleagues in Spain (resp. Italy). The lower Ne values from Mattioni et al. (2008) may be due to their sample size (26 per sampled site) mostly smaller than the other studies.
4.3 No differentiation between chestnut types and a redundant diversity among sampling regions
The absence of significant genetic structure between forest and cultivated stands and a Fct close to zero between both chestnut types indicate common ancestors, similar to other studies (e.g., Pereira-Lorenzo et al. 2019). This result was not completely unexpected given that chestnut is an outcrossing species and that gene flow between forest and cultivated stands is known to occur, together with changes in usage over time and in certain practices such as forests being used as a source of seedlings for rootstock, good quality fruits as a source of seedlings to plant forests, peasant woods in Limousin (personal communication), and “instant domestication” (Pereira-Lorenzo et al. 2019).
Moreover, the high variance found by the AMOVA within sampling regions implies that each sampling region hosts a substantial diversity, mostly shared with the other sampling regions. Such a sharing of genetic diversity between sampling regions confirms the interpretation as the result of human exchanges (Bruneton-Governatori 1999; Conedera et al. 2016; Krebs et al. 2019; Pitte 1986).
Sharing genetic diversity between our sampling regions should also ensure a better backup of this diversity, as long as information about landraces is shared among stakeholders in the different sampling regions. In situ sampling revealed that many landraces are multiclonal. This source of diversity and hence of potential adaptation argues in favor of not reducing a landrace to one arbitrary clone. Even clones should be carefully evaluated, as morphological differences between individual trees attributed to the same clones were reported during our field trips, as has been the case in other species (Cipriani et al. 2010). This result is particularly timely knowing that chestnut valuation tends to be based on heritage, with significance and quality marks associated to local landraces (e.g., AOC Châtaigne d’Ardèche, AOC Farine de châtaigne Corse – Farina castagnina corsa, Label rouge Marron du Périgord). Hasty conclusions coming from a narrow sampling could provide arguments of authority to justify that certain landraces are “local” of the particular area under study. However, even if a landrace has been cultivated for centuries in a particular place, this may also be the case elsewhere. Therefore, one might rightfully ask whether the quality of local chestnut comes from its locality. For crops like chestnut, usage and practices may be at least as important as genetics to give value to chestnut for growers and consumers (Dupré 2002, 2005; Martín et al. 2017).
4.4 Two main genetic clusters
All methods identified a clear genetic structure in all data sets, with at least two clusters. In terms of the number of clusters, the criterion applied to the STRUCTURE results provided the most clear-cut signal; we hence based our analysis on these results. Moreover, the results with less SSRs but more individuals (10Unik data set) were coherent with the results with more SSRs but less individuals (18Unik data set). Even though several articles showed limits to STRUCTURE (e.g., Kalinowski, 2011; Puechmaille 2016) or to the way it can be used (Wang, 2017), a full comparison with the two other methods we tested based on simulations is out of the scope of this article. Still, it is important to note that, at the number of clusters determined by STRUCTURE (K = 2 for 18Unik and K = 6 for 10Unik), the two other methods, DAPC and SNMF, assigned individuals to clusters similar to STRUCTURE. Moreover, even though the SNMF model is closer to the STRUCTURE model than the model behind DAPC (K-means), its assignments of individuals to clusters were less differentiated, as shown by the “clusteredness” statistic.
The two main clusters identified in the French sample separate most trees from Corsica, Var, and Ardèche (RPP1) from those of the other sampling regions (RPP2). In terms of genetic diversity as measured by the effective number of alleles, Ne, both clusters display values (3.27 and 3.04) similar to each other and to those calculated per sampling region (see above). Nevertheless, beyond the structuring in two main clusters, a high admixture remains among sampling regions. Indeed, most of the genetic variance from the AMOVA (> 80%) exists within clusters. Mattioni et al. in 2013 (respectively, 2017) also found such a high percentage with 88.43% (respectively, 87.82%). This contrasts sharply with the study of Skender et al. (2017) on chestnut trees sampled in Bosnia-Herzegovina, Croatia, Slovenia, and Italy (Tyrol), who found that only 22.28% of the genetic variance exists within clusters. They interpreted that such a strong genetic differentiation between clusters comes from the fact that their sample contains “populations most likely originated from independent shelter zones, after the last glaciation period”. Hence, the high admixture we observe in our French sample may suggest that the current chestnut trees in France originate mostly from a single ancestral population. But a detailed investigation in the lines of Krebs et al. (2004, 2019) remains for future work, notably to disentangle the relative importance of the Iberian and Italian refuges with respect to current chestnut in France.
4.5 Future outlook of European genetic structure of chestnut
The genetic structure inferred from our samples as two main clusters did not completely match the sampling regions. This result was also expected for a continuously dispersed species affected by human management like European chestnut. Moreover, an admixed genetic structure was consistent with the known patterns of divergence and distribution of chestnut (Mattioni et al. 2017), combined with evidence from fossil pollen of several tree species suggesting that chestnut populations originating from Italy or the Balkans spread into the Iberian Peninsula from the north (Grivet and Petit 2003; Petit 2003).
In the EU database (2017) and in Pereira-Lorenzo et al. (2019), “Luguesa” was classified with the Italian group of cultivars. In our analyses, it was found in the south-eastern cluster (ESM_8C: RPP1 in 10Unik data set with Spanish samples at K = 2) with “Pais,” “Puga,” and “Raigona,” which were originally classified in the Iberian group. The other Spanish cultivars were found in cluster RPP2. This classification of Spanish samples in our two main clusters is not sufficient to conclude on the spontaneous spread of the species from either Spanish or Italian last glacial refugia (Krebs et al. 2019). However, we provide our genotyping data so that (i) an update of the EU database including more French individual is possible by the community of researchers working on this and (ii) approaches in paleoecology (e.g., Krebs et al. 2004 and Krebs et al. 2019) and palaeodistribution modeling (Roces-Díaz et al. 2018) could be performed at the whole European scale in subsequent studies.
Before removal of hybrid individuals, the Basque sampling region was represented in the 10 (respectively, 18) data set by 119 (respectively, 10) successfully genotyped non-repeated individuals. This high number of admixed individuals is an important feature of the actual chestnut forest there, resulting from the long history of interspecific hybridization in this region, which extends on both sides of the border between Spain and France (Pereira-Lorenzo et al. 2017). It is further substantiated by the high prevalence of trees tolerant to ink disease, as found through artificial inoculation experiments within seedlots we have genotyped (Robin et al., in preparation).
5 Conclusion
In conclusion, this study revealed the genetic diversity and structure of French forest and cultivated chestnut across most of its range. We showed moderate diversity redundancy between sampling regions and a weak genetic structure. Based on external knowledge, the influence of human activity is the most probable explanation for this finding. Two main clusters were found, one in Corsica and the south-east of France. This confirms existing historical knowledge on land use changes, the movement of landraces, and “instant domestication” landraces. We believe our work provides useful information for conservation planning purposes and for cooperation between chestnut non-profit associations and groups of growers interested in landrace conservation and diffusion.
Data availability
The data sets analyzed during the current study are available in the Data.Inrae repository https://doi.org/10.15454/E2DFNJ.
References
Adamack AT, Gruber B (2014) PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5:384–387. https://doi.org/10.1111/2041-210X.12158
Aumeeruddy-Thomas Y, Therville C, Lemarchand C, Lauriac A, Richard F (2012) Resilience of sweet chestnut and truffle holm-oak rural forests in Languedoc-Roussillon, France: roles of social-ecological legacies, domestication, and innovations. Ecol Soc 17:12. https://doi.org/10.5751/ES-04750-170212
Beccaro GL, Torello-Marinoni D, Binelli G, Donno D, Boccacci P, Botta R, Cerutti AK, Conedera M (2012) Insights in the chestnut genetic diversity in Canton Ticino (Southern Switzerland). Silvae Genet 61:292–300
Beghè D, Ganino T, Dall’Asta C, Silvanini A, Cirlini M, Fabbri A (2013) Identification and characterization of ancient Italian chestnut using nuclear microsatellite markers. Sci Hortic 164:50–57. https://doi.org/10.1016/j.scienta.2013.09.009
Bouffartigue C, Debille S, Cabrer AMR, et al (2020) Two main genetic clusters with high admixture between forest and cultivated chestnut (Castanea sativa Mill.) in France. [dataset]. V1. Data Inrae repository. https://doi.org/10.15454/E2DFNJ
Brookfield JFY (1996) A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5:453–455
Bruneton-Governatori A (1999) Le pain de bois: Ethnohistoire de la châtaigne et du châtaignier, Lacour. Nîmes
Buck EJ, Hadonou M, James CJ, Blakesley D, Russell K (2003) Isolation and characterization of polymorphic microsatellites in European chestnut (Castanea sativa Mill.). Mol Ecol Notes 3:239–241. https://doi.org/10.1046/j.1471-8286.2003.00410.x
Chessel D, Dufour AB, Thioulouse J (2004) The ade4 package - I: one-table methods. R news 4:5–10
Cipriani G, Spadotto A, Jurman I, di Gaspero G, Crespan M, Meneghetti S, Frare E, Vignani R, Cresti M, Morgante M, Pezzotti M, Pe E, Policriti A, Testolin R (2010) The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture amongst varieties of different geographic origin. Theor Appl Genet 121:1569–1585. https://doi.org/10.1007/s00122-010-1411-9
Conedera M, Tinner W, Krebs P, et al (2016) Castanea sativa in Europe: distribution, habitat, usage and threats
Dupré L (2002) Du marron à la châtaigne d’Ardèche. La relance d’un produit régional. Éditions du CTHS
Dupré L (2005) Classer et nommer les fruits du châtaignier ou la construction d’un lien à la nature. Nat Sci Sociétés 13:395–402. https://doi.org/10.1051/nss:2005060
Eriksson G, Pliura A, Fernández-López J et al (2005) Management of genetic resources of the multi-purpose tree species Castanea sativa Mill. In: Horticulturae A (ed) International Society for Horticultural Science (ISHS). Leuven, Belgium, pp 373–386
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Smouse PE (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data, p 13
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies, p 22
FAO (2018) Faostat data for chestnut. http://www.fao.org/faostat/.
FAQ List for Arlequin 2000 FAQ List for Arlequin 2.000. http://cmpg.unibe.ch/software/arlequin/software/2.000/doc/faq/faqlist.htm#negative%20variance%20components. Accessed 31 May 2019
Fernández-Cruz J, Fernández-López J (2016) Genetic structure of wild sweet chestnut (Castanea sativa Mill.) populations in northwest of Spain and their differences with other European stands. Conserv Genet 17:949–967. https://doi.org/10.1007/s10592-016-0835-4
Fernández-López J, Fernández-Cruz J (2015) Identification of traditional Galician sweet chestnut varieties using ethnographic and nuclear microsatellite data. Tree Genet Genomes 11. https://doi.org/10.1007/s11295-015-0934-2
FranceAgriMer (2017) chiffres clés Filière Fruits et Légumes 2016
Frascaria N, Blaise S, Guittet J, Lefranc M (1991) Analysis of the spatial genotype distribution in a small chestnut tree population (Castanea sativa MILL.). Spatial autocorrelation and F-statistics. In: Fineshi S, Malvoti ME, Cannata F, HATTEMER HH (eds) Biochemical markers in the population genetics of forest trees. S P B Academic Publ, The Hague, p 219
Frascaria N, Chanson B, Thibaut B, Lefranc M (1992) Gene diversity and wood quality characteristics in chestnut (Castanea sativa Mill.). Ann Sci For 49:49–62. https://doi.org/10.1051/forest:19920105
Frascaria N, Lefranc M (1992) Chestnut trade - a new aspect of the differentiation of chestnut tree (Castanea sativa MILL.) populations in France. Ann Sci For 49:75–79. https://doi.org/10.1051/forest:19920107
Frichot E, Mathieu F, Trouillon T, Bouchard G, François O (2014) Fast and efficient estimation of individual ancestry coefficients. Genetics 196:973–983. https://doi.org/10.1534/genetics.113.160572
Gibson DJ (2014) Methods in comparative plant population ecology. Oxford University Press
Gobbin D, Hohl L, Conza L, Jermini M, Gessler C, Conedera M (2007) Microsatellite-based characterization of the Castanea sativa cultivar heritage of southern Switzerland. Genome 50:1089–1103. https://doi.org/10.1139/G07-086
Goodman SN, Fanelli D, Ioannidis JPA (2016) What does research reproducibility mean? Sci Transl Med 8:341ps12–341ps12. https://doi.org/10.1126/scitranslmed.aaf5027
Goudet J (2005) Hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186. https://doi.org/10.1111/j.1471-8286.2004.00828.x
Grivet D, Petit RJ (2003) Chloroplast DNA phylogeography of the hornbeam in Europe: evidence for a bottleneck at the outset of postglacial colonization, p 10
Guichoux E, Lagache L, Wagner S et al (2011) Current trends in microsatellite genotyping: trends in microsatellite genotyping. Mol Ecol Resour 11:591–611. https://doi.org/10.1111/j.1755-0998.2011.03014.x
Harris SA, Robinson JP, Juniper BE (2002) Genetic clues to the origin of the apple. Trends Genet 18:426–430. https://doi.org/10.1016/S0168-9525(02)02689-6
IGN (2007) BD Forêt® V2. http://professionnels.ign.fr/bdforet.
IGN (2016) RPG. http://professionnels.ign.fr/rpg#tab-1
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. https://doi.org/10.1093/bioinformatics/btm233
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026. https://doi.org/10.1111/j.1365-294X.2008.03887.x
Kalinowski ST (2011) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity 106:625–632. https://doi.org/10.1038/hdy.2010.95
Kampfer S, Lexer C, Glössl J, Steinkellner H (1998) Characterization of (GA)n microsatellite loci from Quercus Robur. Hereditas 129:183–186. https://doi.org/10.1111/j.1601-5223.1998.00183.x
Kamvar ZN, Brooks JC, Grünwald NJ (2015) Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front Genet 6:10. https://doi.org/10.3389/fgene.2015.00208
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. https://doi.org/10.7717/peerj.281
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725–738
Krebs P, Conedera M, Pradella M et al (2004) Quaternary refugia of the sweet chestnut (Castanea sativa Mill.): an extended palynological approach. In: Vegetation history and Archaeobotany, vol 13, p 18. https://doi.org/10.1007/s00334-004-0041-z
Krebs P, Pezzatti GB, Beffa G, Tinner W, Conedera M (2019) Revising the sweet chestnut (Castanea sativa Mill.) refugia history of the last glacial period with extended pollen and macrofossil evidence. Quat Sci Rev 206:111–128. https://doi.org/10.1016/j.quascirev.2019.01.002
Lassois L, Denancé C, Ravon E, Guyader A, Guisnel R, Hibrand-Saint-Oyant L, Poncet C, Lasserre-Zuber P, Feugey L, Durel CE (2016) Genetic diversity, population structure, parentage analysis, and construction of core collections in the French apple germplasm based on SSR markers. Plant Mol Biol Report 34:827–844. https://doi.org/10.1007/s11105-015-0966-7
Lusini I, Velichkov I, Pollegioni P, Chiocchini F, Hinkov G, Zlatanov T, Cherubini M, Mattioni C (2013) Estimating the genetic diversity and spatial structure of Bulgarian Castanea sativa populations by SSRs: implications for conservation. Conserv Genet 15:283–293. https://doi.org/10.1007/s10592-013-0537-0
Marinoni D, Akkak A, Bounous G, Edwards KJ, Botta R (2003) Development and characterization of microsatellite markers in Castanea sativa (Mill.). Mol Breed 11:127–136. https://doi.org/10.1023/A:1022456013692
Martín MA, Mattioni C, Cherubini M, Taurchini D, Villani F (2010a) Genetic characterisation of traditional chestnut varieties in Italy using microsatellites (simple sequence repeats) markers. Ann Appl Biol 157:37–44. https://doi.org/10.1111/j.1744-7348.2010.00407.x
Martín MA, Mattioni C, Cherubini M et al (2010b) Genetic diversity in European chestnut populations by means of genomic and genic microsatellite markers. Tree Genet Genomes 6:735–744. https://doi.org/10.1007/s11295-010-0287-9
Martín MA, Monedero E, Martín LM (2017) Genetic monitoring of traditional chestnut orchards reveals a complex genetic structure. Ann For Sci 74:13. https://doi.org/10.1007/s13595-016-0610-1
Mattioni C, Cherubini M, Micheli E, Villani F, Bucci G (2008) Role of domestication in shaping Castanea sativa. Tree Genet Genomes 4:563–574. https://doi.org/10.1007/s11295-008-0132-6
Mattioni C, Martín MA, Chiocchini F et al (2017) Landscape genetics structure of European sweet chestnut (Castanea sativa Mill): indications for conservation priorities. Tree Genet Genomes 13:39. https://doi.org/10.1007/s11295-017-1123-2
Mattioni C, Martín MA, Pollegioni P et al (2013) Microsatellite markers reveal a strong geographical structure in European populations of Castanea sativa (Fagaceae): evidence for multiple glacial refugia. Am J Bot 100:951–961. https://doi.org/10.3732/ajb.1200194
Mellano MG, Beccaro GL, Donno D, Marinoni DT, Boccacci P, Canterino S, Cerutti AK, Bounous G (2012) Castanea spp. biodiversity conservation: collection and characterization of the genetic diversity of an endangered species. Genet Resour Crop Evol 59:1727–1741. https://doi.org/10.1007/s10722-012-9794-x
Mellano MG, Torello-Marinoni D, Boccacci P et al (2018) Ex situ conservation and characterization of the genetic diversity of Castanea spp. In: Acta Horticulturae, pp 1–6. https://doi.org/10.17660/ActaHortic.2018.1220.1
Míguez-Soto B, Fernández-Cruz J, Fernández-López J (2019) Mediterranean and Northern Iberian gene pools of wild Castanea sativa Mill. are two differentiated ecotypes originated under natural divergent selection. PLoS One 14:e0211315. https://doi.org/10.1371/journal.pone.0211315
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Paradis E (2010) Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420. https://doi.org/10.1093/bioinformatics/btp696
Pereira-Lorenzo S, Costa RML, Ramos-Cabrer AM, Ribeiro CAM, da Silva MFS, Manzano G, Barreneche T (2010) Variation in grafted European chestnut and hybrids by microsatellites reveals two main origins in the Iberian Peninsula. Tree Genet Genomes 6:701–715. https://doi.org/10.1007/s11295-010-0285-y
Pereira-Lorenzo S, Fernandez-Lopez J (1997) Description of 80 cultivars and 36 clonal selections of chestnut (Castanea sativa Mill.) from Northwestern Spain. Fruit Variet J 51:13–27
Pereira-Lorenzo S, Costa R, Anagnostakis S et al (2016) Interspecific hybridization of chestnut. In: Mason AS (ed) Polyploidy and hybridization for crop improvement. CRC Press, Boca Raton, pp 377–407. https://doi.org/10.1201/9781315369259-16
Pereira-Lorenzo S, Ramos-Cabrer A, Barreneche T et al (2017) Database of European chestnut cultivars and definition of a core collection using simple sequence repeats. Tree Genet Genomes 13:5. https://doi.org/10.1007/s11295-017-1197-x
Pereira-Lorenzo S, Ramos-Cabrer AM, Barreneche T, Mattioni C, Villani F, Díaz-Hernández B, Martín LM, Robles-Loma A, Cáceres Y, Martín A (2019) Instant domestication process of European chestnut cultivars. Ann Appl Biol 174:74–85. https://doi.org/10.1111/aab.12474
Petit RJ (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565. https://doi.org/10.1126/science.1083264
Pitte J-R (1986) Terres de Castanide. In: Hommes et paysages du Châtaignier de l’Antiquité à nos jours. Fayard, Paris
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627. https://doi.org/10.1111/1755-0998.12512
Quintana J, Contreras A, Merino I et al (2014) Genetic characterization of chestnut (Castanea sativa Mill.) orchards and traditional nut varieties in El Bierzo, a glacial refuge and major cultivation site in northwestern Spain. In: Tree Genetics & Genomes, p 11. https://doi.org/10.1007/s11295-014-0826-x
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Roces-Díaz JV, Jiménez-Alfaro B, Chytrý M, Díaz-Varela ER, Álvarez-Álvarez P (2018) Glacial refugia and mid-Holocene expansion delineate the current distribution of Castanea sativa in Europe. Palaeogeogr Palaeoclimatol Palaeoecol 491:152–160. https://doi.org/10.1016/j.palaeo.2017.12.004
Rosenberg NA, Mahajan S, Ramachandran S et al (2005) Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet 1:660–671. https://doi.org/10.1371/journal.pgen.0010070
Sauvezon R, Sauvezon A, Sunt C (2000) Châtaignes et Châtaigniers. Edisud
Skender A, Kurtovic M, Pojskic N, Kalamujic-Stroil B, Hadziabulic S, Gasi F (2017) Genetic structure and diversity of European chestnut (Castanea sativa Mill.) populations in western Balkans: on a crossroad between east and west. Genetika 49:613–626. https://doi.org/10.2298/GENSR1702613S
Steinkellner H, Fluch S, Turetschek E et al (1997) Identification and characterization of (GA/CT)n- microsatellite loci from Quercus petraea. Plant Mol Biol 33:1093–1096. https://doi.org/10.1023/A:1005736722794
Villa TCC, Maxted N, Scholten M, Ford-Lloyd B (2005) Defining and identifying crop landraces. Plant genetic resources: characterization and utilization 3:373–384. https://doi.org/10.1079/PGR200591
Wang J (2017) The computer program STRUCTURE for assigning individuals to populations: easy to use but easier to misuse. Mol Ecol Resour 17:981–990. https://doi.org/10.1111/1755-0998.12650
Acknowledgments
This paper is part of the PhD of the first author who is grateful of her supervisors Laurent Hazard and Nathalie Couix of the INRAE, AGIR, and Timothée Flutre, INRAE, Le Moulon.
We would like to thank the local partners who introduced the first author to the chestnut, helped in sampling, and shared their knowledge, in particular are the members of ACRC, Châtaigne des Pyrénées, Rénova, SPCV, and Paysans du Rance.
We would like to thank the people who provided access to forest samples or sampling regions: in Corsica, GRPTCMC and INRAE Corsica; in Limousin, Croqueurs de pommes du Limousin and the Parc Naturel Régional Perigord-Limousin; and in Ardèche and Eastern Occitanie, Chambre départementale d’agriculture d’Ardèche (CDA Ardèche), Syndicat des producteurs de châtaigne d’Ardèche (SDCA), and Chambre régionale d’agriculture d’Occitanie (CRA Occitanie), and also other public and private landowners who granted us access to their forest stands in Brittany, Gard, Aveyron, Basque Country, and Cantal.
We are very grateful to advisors and partners taking part to other aspects of our work on chestnut such as Invenio, CTIFL, INRA, Biogeco, INRA, and Biologie du Fruit et Pathologie, CIF-Lourizan (Spain) and the members of the national workgroup on chestnut forestry set up by the National Center for Private Forest owners (CNPF) and particularly its chairman René Lempire as well as Jean Lemaire and Sabine Girard.
We would also like to thank FCBA colleagues involved in sampling.
The authors thank the anonymous reviewers for critically reading the manuscript and suggesting substantial improvements.
Funding
A grant of the Fondation de France and a regional research program for and on rural development (PSDR4-Occitanie, France) fund the PhD of the first author.
In 2016/2017, local partners (Rénova, ACRC, and Châtaigne des Pyrénées) collected subsidies from the Conservatory of the Regional Biological Patrimony (CPBR) for the genotyping.
The FCBA project on forest samples was funded by the Conseil Régional de Nouvelle Aquitaine through the “sélection châtaignier à bois” project (2015–2016) contract number 15007198-046 and grant “FEDER-FSA 2014-2020 Dossier 47010” and “sélection châtaignier” project grant n°16008302-043 and “FEDER-FSE 2014-2020 – AXE 1 n° 3296610”. The two FEDER grant parts are funds obtained from European Union by the Conseil Régional de Nouvelle Aquitaine. We thank the members of the FCBA External Advisory Board on Forestry and members of the National Center for private forest owners for their help on setting up and managing the abovementioned projects.
The Xylobiotech platform is funded through the Equipe Xyloforest project (grant ANR-10-EQPX-16).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest
Additional information
Handling Editor: Ricardo Alia
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contribution of the co-authors
Conceptualization, Cathy Bouffartigue, Timothée Flutre, and Luc Harvengt; Methodology, Cathy Bouffartigue, Sandrine Debille, Ana Ramos Cabrer, Timothée Flutre, and Luc Harvengt; Software, Cathy Bouffartigue and Timothée Flutre; Validation, Cathy Bouffartigue and Timothée Flutre; Formal Analysis, Cathy Bouffartigue, Ana Ramos Cabrer, Santiago Pereira-Lorenzo, and Timothée Flutre; Investigation, Cathy Bouffartigue, Sandrine Debille, Olivier Fabreguette, and Ana Ramos Cabrer; Resources, Cathy Bouffartigue, Sandrine Debille, Olivier Fabreguette, Ana Ramos Cabrer, Santiago Pereira-Lorenzo, and Luc Harvengt; Data curation, Cathy Bouffartigue and Sandrine Debille; Writing (original draft), Cathy Bouffartigue and Timothée Flutre; Writing (review and editing), Cathy Bouffartigue, Sandrine Debille, Ana Ramos Cabrer, Santiago Pereira Lorenzo, Timothée Flutre, and Luc Harvengt; Visualization, Cathy Bouffartigue; Reproducible analysis, Timothée Flutre and Cathy Bouffartigue; Supervision, Timothée Flutre and Luc Harvengt; Project administration, Cathy Bouffartigue, Timothée Flutre, and Luc Harvengt; Funding acquisition, Cathy Bouffartigue and Luc Harvengt.
Rights and permissions
About this article
Cite this article
Bouffartigue, C., Debille, S., Fabreguettes, O. et al. Two main genetic clusters with high admixture between forest and cultivated chestnut (Castanea sativa Mill.) in France. Annals of Forest Science 77, 74 (2020). https://doi.org/10.1007/s13595-020-00982-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13595-020-00982-w