Abstract
Key message
The dataset provides an exhaustive tree inventory with forest mensuration and spatial location carried out in 54 plots sampled in 45- to 55-year-old black pine plantations, located in two areas of Tuscany (central Italy). Forest mensuration includes horizontal and vertical structure measurements and a total of 4171 trees were geo-referenced. The most abundant species was the black pine, Pinus nigra spp. laricio , for which a total of 3631 trees were observed. The dataset was collected as part of the SelPiBio LIFE project (LIFE13 BIO/IT/000282). Dataset access at http://doi.org/10.5281/zenodo.438681 . Associated metadata available at https://metadata-afs.nancy.inra.fr/geonetwork/apps/georchestra/?uuid=73591027-0f1e-40a3-95d0-b614517c1290&hl=eng .
Context
The main aim of the SelPiBio LIFE project (www.selpibio.eu) is to demonstrate the effects of two thinning regimes, selective and from below, on soil biodiversity in young black pine stands. The spatial structure of forests and the relationships between trees are a good proxy of overall biodiversity level. Spatial datasets with geo referenced trees and related mensurational data represent the highest level of information for forest inventories and research activities.
Aims
This dataset has been developed during the A2 Action (Assessment of structural and mensurational parameters of the forest stands and the dead wood) of the project, to record the main mensurational parameters of the studied black pine stands. A tree-level database was compiled to describe the vertical and horizontal structure of 54 monitoring plots before the application of the silvicultural treatment.
Methods
In addition to classical in-field measurements (e.g. diameters at breast height, total height of the tree, crown depth etc.), all trees were georeferenced by means of polar coordinates collected from the centre of each monitoring plot, including crown projection on the ground, described with eight points. Then, a polynomial spline function was fitted across the recorded data to obtain a convex polygon and to calculate crown area and crown perimeter of each measured tree in GIS environment.
Results
A polygonal ESRI shapefile in ETRS89/UTM32N reference system (EPSG: 25832) with 4171 records representing the crown projections on the ground of each measured tree with all the mensurational parameters included into the attribute table. The database is freely available under the Creative Commons Attribution-NonCommercial 4. 182 0 License.
Conclusion
With this database, a wide range of forestry-related indices could be easily calculated, including geostatistical analysis and autocorrelation functions, to compare Italian artificial black pine stands with other studied forests.
Similar content being viewed by others
1 Background
This dataset has been developed during the A2 Action of the SelPiBio LIFE project (www.selpibio.eu/en). The main aim of this project is to demonstrate the effects of two thinning regimes, selective and from below, on soil biodiversity in young black pine stands. Indeed, even if its efficacy on black pine plantations (Pinus nigra spp. laricio) has been already discussed in literature (Cantiani et al. 2010), selective thinning is still an uncommon silvicultural treatment in such forests of Italy.
To evaluate the effect of thinning on pine plantations, 18 monitoring areas of 1 ha each (hereafter “sector”) were designed and realized across two study areas in Tuscany (central Italy), the Pratomagno mountain chain and the Monte Amiata. In each study area (hereafter “zone”), 27 circular plots of 15 m of radius were geo-referenced (three for each sector). Between 2015 and early 2016, all included trees were measured, to characterize both horizontal and vertical structure of plantations, highly relevant for forest management purposes (Latham et al. 1998; Cantiani et al. 2010; Pretzsch et al. 2016). The database was finalized to study the effects of the two tested thinning approaches in modifying both the forest vertical and horizontal stand structure, proxy of the overall biodiversity level (Neumann and Starlinger 2001).
2 Methods
2.1 Study design and sampling
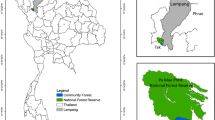
The spatial distribution of the sampling scheme was studied during the last quarter of 2014. In this first period, the forest coverage of the two zones (Monte Amiata and Pratomagno) was analysed with most updated orthophotos provided by the Regional administration of Tuscany as Web Map Service (WMS). Then, the two selected study areas (approximately 20 ha each) were divided into nine squares of 1 ha each. Afterwards, three circular plots of 15 m of radius were randomly sampled inside each square for a total of 54 sampling units (plots). Then, all data were charged on a GPS system and materialized on the field. Due to GPS uncertainties, the spatial distribution of the forest (some gaps were detected on the canopy coverage form aerial photos) and the random procedure, some plots were overlapping. Anyway, this issue did not represent an obstacle for our sampling scheme; indeed, each plot was maintained and analysed separately. Common trees were measured just once and replicated in the second plot. Moreover, in such cases, one of the two centres was always geo-referenced using the polar coordinates (as explained below in the next subsection for trees) collected from the other centre of the plot. In this way, GPS uncertainty was charged just on one centre, while the second one and all the related trees (and especially those in common between the two circular plots) were co-registered with relative coordinates and centrimetrical precision around the centimetre. The spatial distribution of 1-ha sectors and 15-m-radius circular plots are reported in Fig. 1 for the two zones.
2.2 Field measurements
The survey campaign was completed between 2015 and early 2016. All trees included into the 54 circular plots were fully measured, as well as all the external trees whose crown projection fell inside the plot. This was done to avoid underestimation of the canopy cover. All trees inside the circular plot were numbered progressively (from 1 to n in each plot) while a “99” was added before the progressive number in case of external ones (e.g. while the trees nr. 1, 7 or 12 were inside the plot, the trees nr. 991, 997 or 9912 were laying outside). Then, a unique ID number was given to each tree as concatenation between the zone code (Monte Amiata = 1, Pratomagno = 2), the monitoring sector (from 1 to 9), the plot number (from 1 to 3) and the tree number (e.g. the ID 23112 correspond to the Pratomagno zone, monitoring sector 3, plot 1 and the tree number 12).
The spatial structure of the plots was recorded using a custom version of the FieldMap® technology composed by a Trupulse360B connected to a portable tablet with Bluetooth connection. In this handmade version, manually developed following the instruction of the user guide of Trupulse360B, a Visual BASIC script was written to upload the horizontal distance and the azimut form the Trupulse360B directly on a spreadsheet.
During in-field works, each tree was geo-referenced using polar coordinate system (horizontal distance from the centre of the circular plot and degrees from the north), subsequently converted into metric planar coordinates (X, Y). After correcting the obtained horizontal distance by adding the radius of each tree, the conversion procedure was performed by applying one of the basic rules of right triangle geometry. Following Fig. 2, suppose α to be the measured angle from the north (increasing clockwise) and the hypotenuse i to be the horizontal distance between the centre of plot A and the tree T to be geo-referenced. Then, the two legs of the right triangle (c 1 and c 2) can easily be calculated using the following formulas:
where c 1 corresponds to the Y coordinate and c 2 corresponds to the X coordinate, respectively, and both the resulting coordinates are expressed in metres. Then, adding the relative X and Y coordinates (in metres) to the geographic position of the central point (in a metric system e.g. UTM or LAEA), each tree can be easily geo-referenced in the same reference system. Using this approach, only the central point of (some) plots was geo-referenced using the GPS system, avoiding many spatial biases and improving geometric accuracy and precision of the database. Similarly, also the crown projection on the ground was recorded in the same way. Eight measurements (points) were used to project the crown on the ground. Then, a polynomial spline function across the eight points was fitted to obtain a convex polygon.
Once all the trees inside all plots were measured, the thinning was applied to the whole sector. All the trees to be removed were marked on the field and recorded on the final database; then, the treatment was applied (except control sectors) between spring and summer 2016.
2.3 Dataset content
To characterize each tree, a tree-level database was compiled with the following information: (i) species of the tree (SP), (ii) rank (dominant—intermediate—dominated—dead—snag), (iii) diameter at breast height (DBH), (iv) total height of the tree (HT), (v) height of maximum crown width (HMCW), (vi) height of the first living branch (HFLB) and (vii) living whorl number (LWN, Cantiani and Chiavetta 2015; Marchi et al. 2017). In the end, an ESRI shapefile was created including all the collected information (DBH, RANK, LWN, etc.) except X and Y coordinates of the eight crown points that have been substituted with polygons created with spline interpolation (with 100 points), using a script implemented in R language (R CoreTeam 2017). Finally, all the data for the two areas were merged in a unique ESRI shapefile in ETRS89/UTM32N reference system (EPSG 25832). In this database, each tree is unequivocally identified by its ID number and represented by its crown projection with all the associated information included into the .dbf file. If necessary, the exact stem position can be easily extracted from the .dbf file to create, for instance, a new spatial point object. The last column of the database, named “REMOVED”, has been included to simulate the applied thinning scheme (“TREATMENT” column) to each circular plot and to derive all the quantitative statistics of the two treatments. Summary statistics of the collected data before the treatment are reported in Table 1.
3 Access to data and metadata description
The database can be downloaded using the following reference and doi: Cantiani P, Marchi M (2017). A spatial dataset of forest mensuration collected in black pine plantations in central Italy [Data set]. V1. CREA. http://doi.org/10.5281/zenodo.438681 under the Creative Commons Attribution-NonCommercial 4.0 License. Associated metadata available at https://metadata-afs.nancy.inra.fr/geonetwork/apps/georchestra/?uuid=73591027-0f1e-40a3-95d0-b614517c1290&hl=eng.
4 Technical validation
The database includes 4171 trees (lines) and 21 columns. Each tree has been carefully checked before publication, avoiding misspelling or errors. The standardization of records (e.g. correction of the species code or the rank if upper case instead of lower case) has been prevented not only by involving expert field technicians and researchers but also in post-processing steps by means of filters and cross-check procedures (the database is case-sensitive). The database has been compiled in metric units using the dot as decimal value.
Before fitting the spline function, all crown projections were controlled to prevent artefacts due to missing records or sampled points too close to each other. In fact, in this last case, a ribbon was very likely to be created using the spline function, bringing the calculation process to failure due to an invalid polygonal form. The information about thinning (REMOVED column) was firstly stored on the database and afterwards validated after the treatment. This was done to control the harvesting procedure and to avoid discrepancy between real situation and database (e.g. uncut trees or damages to alive ones).
5 Reuse potential and limits
Even if characterized by small plots, the sampling design and the high number of repetitions in each zone and treatment may allow the calculation of a wide number of forestry-related indices, useful to describe ecological dynamics (Neumann and Starlinger 2001; Corona et al. 2002; Chiavetta et al. 2016). Volumes could be easily calculated with ForIT package of R (Puletti et al. 2017), an object-oriented tool, which implements all volume equations of the last Italian National Forest Inventory, available on public R CRAN repository and also GitHub. This ensemble will also allow further comparisons with other European artificial pinewoods as well as native zones for the widely distributed P. nigra spp. (Ciancio et al. 2006; Amodei et al. 2012; Marchi et al. 2015; Enescu et al. 2016).
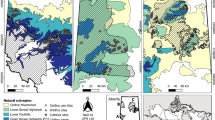
Thanks to the adopted structure (ESRI shapefile), also geostatistical analysis could be easily performed, e.g. to study the autocorrelation function or to estimate mark variograms (Stoyan and Walder 2000; Pommerening and Särkkä 2013) following the principles of the spatial point pattern analysis (Baddeley and Turner 2005; Baddeley et al. 2015) or replicated pattern analysis (Ramon et al. 2016). In addition, the shapefile may lead to more flexibility to further research activities. In such case, researchers could choose to load it into a pure GIS or a statistical program such as R environment, using for example the rgdal package (Bivand et al. 2015), as well as to extract the database as spreadsheet using just the .dbf file. A spatial representation of a test plot with QGIS is reported in Fig. 3.
In the future, further connections with soil biodiversity data, trees’ response to the applied thinning system or other data from the SelPiBio LIFE project may enrich the provided source.
References
Amodei T, Guibal F, Fady B (2012) Relationships between climate and radial growth in black pine (Pinus nigra Arnold ssp. salzmannii (Dunal) Franco) from the south of France. Ann For Sci 70:41–47. doi:10.1007/s13595-012-0237-9
Baddeley A, Turner R (2005) Spatstat: an R package for analyzing spatial point patterns. J Stat Softw 12:1–42
Baddeley A, Rubak E, Turner R (2015) Spatial point patterns: methodology and applications with R. Series: Chapman & Hall/CRC Interdisciplinary Statistics. Ed CRC press. ISBN 9781482210200, 810 pp
Bivand R, Keitt T, Rowlingson B (2015) rgdal: bindings for the Geospatial Data Abstraction Library
Cantiani P, Chiavetta U (2015) Estimating the mechanical stability of Pinus nigra Arn. using an alternative approach across several plantations in central Italy. iForest - Biogeosciences For 8:846–852. doi:10.3832/ifor1300-007
Cantiani P, Marchi M (2017) A spatial dataset of forest mensuration collected in black pine plantations in central Italy [Data set]. V1. CREA. doi:10.5281/zenodo.438681
Cantiani P, Plutino M, Amorini E (2010) Effects of silvicultural treatment on the stability of black pine plantations. Ann Silvic Res 36:49–58
Chiavetta U, Skudnik M, Becagli C, Bertini G, Ferretti F, Cantiani P, Di Salvatore U, Fabbio G (2016) Diversity of structure through silviculture diversity of structure through silviculture. Ital J Agron 11:18–22
Ciancio O, Iovino F, Menguzzato G, Nicolaci A, Nocentini S (2006) Structure and growth of a small group selection forest of calabrian pine in southern Italy: a hypothesis for continuous cover forestry based on traditional silviculture. For Ecol Manag 224:229–234. doi:10.1016/j.foreco.2005.12.057
Corona P, Marziliano PA, Scotti R (2002) Top-down growth modelling: a prototype for poplar plantations in Italy. For Ecol Manag 161:65–73. doi:10.1016/S0378-1127(01)00481-9
Enescu C, de Rigo D, Caudullo G et al (2016) Pinus nigra in Europe: distribution, habitat, usage and threats. In: San-Miguel-Ayanz J, de Rigo D, Caudullo G, Houston Durrant T, Mauri A (eds) European atlas of Forest tree species, pp 126–127
Latham P, Zuuring H, Coble D (1998) A method for quantifying vertical forest structure. For Ecol Manag 104:157–170
Marchi M, Castaldi C, Merlini P, Nocentini S, Ducci F (2015) Stand structure and influence of climate on growth trends of a marginal forest population of. Ann Silvic Res 39:100–110. doi:10.12899/ASR-1066
Marchi M, Chiavetta U, Cantiani P (2017) Assessing the mechanical stability of trees in artificial plantations of Pinus nigra J. F. Arnold using the LWN indicator under different site indexes. Ann Silvic Res 41(1):48–53. doi:10.12899/ASR-1312
Neumann M, Starlinger F (2001) The significance of different indices for stand structure and diversity in forests. For Ecol Manag 145:91–106. doi:10.1016/S0378-1127(00)00577-6
Pommerening A, Särkkä A (2013) What mark variograms tell about spatial plant interactions. Ecol Model 251:64–72. doi:10.1016/j.ecolmodel.2012.12.009
Pretzsch H, del Río M, Schütze G, Ammer C, Annighöfer P, Avdagic A, Barbeito I, Bielak K, Brazaitis G, Coll L, Drössler L, Fabrika M, Forrester DI, Kurylyak V, Löf M, Lombardi F, Matović B, Mohren F, Motta R, den Ouden J, Pach M, Ponette Q, Skrzyszewski J, Sramek V, Sterba H, Svoboda M, Verheyen K, Zlatanov T, Bravo-Oviedo A (2016) Mixing of scots pine (Pinus sylvestris L.) and European beech (Fagus sylvatica L.) enhances structural heterogeneity, and the effect increases with water availability. For Ecol Manag 373:149–166. doi:10.1016/j.foreco.2016.04.043
Puletti N, Castaldi C, Marchi M, Scotti R (2017) ForIT: Functions from the 2nd Italian Forest Inventory (INFC). https://github.com/cran/ForIT/blob/master/R/ForIT-package.R
R CoreTeam (2017) R: a language and environment for statistical computing, Vienna, Austria. URL //www.R-project.org/. Accessed 18 Jan 2017
Ramon P, de la Cruz M, Chacon-Labella J, Escudero A (2016) A new non-parametric method for analyzing replicated point patterns in ecology. Ecography (Cop) 39:1109–1117. doi:10.1111/ecog.01848
Stoyan D, Walder O (2000) On variograms in point process statistics, II: models of markings and ecological interpretation. Biom J 42:171–187. doi:10.1002/(SICI)1521-4036(200005)42:2<171::AID-BIMJ171>3.0.CO;2-L
Acknowledgements
Authors wish to thank Valter Cresti, Maurizio Piovosi, Manuela Plutino, Luca Marchino, Alice Angelini, Mirko Grotti and Giulia Rinaldini from the Research Centre for Forestry and Wood (CREA-FL) for the external assistance during the survey campaign. We are also very grateful to the two anonymous reviewers and the data paper handling editor Marianne Peiffer for the very useful comments on the early draft of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
EU LIFE financial instrument, LIFE13 BIO/IT/000282
Additional information
Handling Editor: Marianne Peiffer
Rights and permissions
About this article
Cite this article
Cantiani, P., Marchi, M. A spatial dataset of forest mensuration collected in black pine plantations in central Italy. Annals of Forest Science 74, 50 (2017). https://doi.org/10.1007/s13595-017-0648-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13595-017-0648-8