Abstract
Melipona bees are remarkable due to the high contrast in heterochromatin amounts, making this group interesting for studying repetitive DNA amplification. Here, we performed the first efforts for the chromosomal localization of different repetitive DNAs in M. (Michmelia) scutellaris and tested for unique or multiple heterochromatin amplification in Melipona subgenera. Our data revealed enrichment of repetitive DNAs in chromosomal heterochromatic arms demonstrated by C0t-DNA and DOP-PCR probe hybridization, although microsatellites and multigene families were located at terminal euchromatic regions. Analysis using C0t-DNA probe from M. scutellaris showed positive hybridization only in Michmelia species, suggesting monophyletic amplification and sharing of heterochromatin sequences between species. However, the subgenus Melikerria, with a high amount of heterochromatin, probably underwent independent heterochromatin amplification or experienced sequence modification.
Similar content being viewed by others
1 Introduction
Eukaryote genomes are composed of the following two types of repetitive DNAs: (i) in tandem arranged elements, including satellite DNAs, microsatellites, minisatellites, and several multigene families, and (ii) scattered elements, which are represented by transposons and retrotransposons (Charlesworth et al. 1994; Nowak 1994, López-Flores and Garrido-Ramos 2012; Martins 2007). The abundance, easy isolation, and facility in chromosome mapping, allowed the frequent use of these sequences as markers to understand chromosomal and genome organization and evolution in most diverse taxa, including some insect groups. However, few studies have been undertaken in bees (Brito et al. 2005; Rocha et al. 2002; Lopes et al. 2014).
Stingless bees (Meliponini) comprise a diverse group of highly eusocial insects mostly occurring in the New World tropics (Michener 2007, 2013; Camargo 2013). They have great ecological importance as pollinators of native and agricultural plants (Heard 1999; Pedro 2014). The genus Melipona comprises at least 50 species distributed from Mexico to Argentina (Michener 2007). Melipona species have been the focus of ecological, behavioral, phylogenetic, and genetic studies, but studies focused on their chromosomes are still scarce (Roubik 2006; Rocha et al. 2007; Ramirez et al. 2010; Tavares et al. 2017).
Until now, Melipona karyotypes had been described for 24 species, displaying a very conserved chromosome number of n = 9 (2n = 18). This was observed in 23 species and only two M. seminigra subspecies showed n = 11, which probably originated from fissions in two chromosomes (revised by Tavares et al. 2017). Interestingly, the heterochromatin amounts and chromosomal distribution are highly divergent in Melipona. According to Rocha and Pompolo (1998), they can be divided into two groups when analyzing by C-banding. One group comprises species with low heterochromatic content (less than 50%) that is primarily distributed in pericentromeric regions or in the short chromosomal arms of their chromosomes (group I). The other group includes species with high heterochromatic content, which occupy almost the entire chromosomes (group II). The heterochromatin in Melipona was studied until now by C-banding and fluorochrome staining providing only general information about distribution and base pair enrichment (reviewed by Rocha et al. 2007).
To better understand the chromosomal distribution of repetitive sequences and their relationship with eu- or heterochromatin in the genus Melipona, we characterized the chromosomes from M. (Michmelia) scutellaris, a species with high heterochromatic content, using distinct probes for repetitive DNAs. Moreover, to test the hypothesis of multiple or unique rounds of heterochromatin amplification and dispersion in Melipona, we isolated the pool of repetitive DNAs (C0t-DNA fraction) from M. scutellaris and used as probe against genomic DNA from five other species belonging to the distinct subgenera and displaying the distinct heterochromatin distribution.
2 Materials and methods
Melipona scutellaris larvae were obtained from apiary colonies at the Bioscience Institute at São Paulo State University, Rio Claro/SP. The mitotic chromosomes were obtained according to Imai et al. (1988) using the cerebral ganglia from post-defective larvae as the source. Slides were prepared by maceration followed by drying on a heat plate (45 °C). For conventional analyses, the slides were stained with 5% Giemsa for chromosome counting and heterochromatin detection was performed according to Sumner (1972).
Genomic DNA was extracted from adults using the phenol/chloroform method (Sambrook and Russel 2001). Multigene families were amplified by polymerase chain reaction (PCR) using specific primers for the 18S rDNA (Cabral-de-Mello et al. 2010) and U2 snDNA (Bueno et al. 2013). Two strategies were used to obtain the repetitive DNA pools: (i) the reassociation kinetics technique (C0t) and (ii) DOP-PCR. The reassociation kinetics technique (C0t) was performed according to Zwich et al. (1997) with modifications. Approximately 8 μg of DNA was fragmented with DNAse (Sigma-Aldrich) for 1 min. The generated fragments were checked by 1% agarose gel electrophoresis. The DNA samples were then denatured at 95 °C for 10 min and incubated on ice for 10 s. Then, the samples were subjected to a reassociation temperature of 65 °C for 10 min. After the samples were incubated at 37 °C for 8 min with S1 nuclease, the reaction was stopped by the addition of liquid nitrogen. The DNA was purified with phenol/chloroform (1:1, v/v) and the results were confirmed by electrophoresis on a 1% agarose gel. For amplification by DOP-PCR, a protocol proposed by Telenius et al. (1992) using the degenerate primer 5′-CCGACTCGACNNNNNNATCTGG was utilized.
The repetitive DNA sequences were labeled with digoxigenin 11-dUTP or biotin 11-dATP by PCR or nick-translation. The seven microsatellite probes were directly labeled with biotin-14 dATP at the 5′-end during their synthesis (Sigma, St Louis, MO, USA). Fluorescent in situ hybridization (FISH) followed the protocol described by Pinkel et al. (1986), with modifications proposed by Cabral-de-Mello et al. (2010), and the probes were detected using anti-digoxigenin-rhodamine (Roche) or Alexa-fluor-488-conjugated streptavidin (Life Technologies). All the preparations were stained with DAPI and mounted with Vectashield mounting medium (Vector, Burlingame, CA, USA). FISH signals were observed using an Olympus BX61 microscope equipped with a fluorescent lamp.
To check whether the pool of repetitive DNAs obtained by C0t-DNA from M. scutellaris is shared with other species from the same genus, we performed dot-blot analysis according to the protocol described by Anjos et al. (2016). The probe was tested against genomic DNA from species with low amount of heterochromatin M. (Eomelipona) bicolor and M. (Melipona) quadrifasciata and with high amount of heterochromatin M. (Melikerria) fasciculata, M. (Michmelia) rufiventris, and M. (Michmelia) seminigra. The genomic DNA of M. (Michmelia) scutellaris was used as positive control.
3 Results and discussion
The 2n = 18 (female) observed in M. scutellaris (Figure 1) was previously reported by Rocha et al. (2007). This condition was also reported in 22 other species from the same genus, with variant diploid numbers only occurring in M. seminigra when considering the A complement. Among Meliponini species including Melipona, the diploid number conservation is recurrent in distinct genera, with slight variations (revised by Tavares et al. 2017).
The mapping of repetitive DNAs is scarce in bees, and FISH mapping using multigene families as a probe in Melipona is restricted to major rDNAs in a few species (Rocha et al. 2002, 2007). A common feature of the Melipona karyotypes is the heteromorphism in size for the chromosome 1, and in M. scutellaris, this chromosome carries 18S rRNA at the terminal region (Figure 1). rDNA loci restricted to one chromosomal pair were also observed in other species of Melipona through FISH. Similar data was observed through silver nitrate staining (Rocha et al. 2002; Rocha 2002). It could indicate a modal pattern in the genus, but a higher number of species should be studied. This pattern is also recurring in other Hymenoptera, such as parasitic and social wasps (Gokhman et al. 2014; Menezes et al. 2013) and ants (Imai et al. 2001). However, reports of multiple 18S rDNA signals were also described in Hymenoptera, as in Partamona (Meliponini) (Brito et al. 2005) and some parasitoid wasps (Paladino et al. 2013; Gokhman et al. 2014). The U2 snDNA was located in pair 2 at the terminal region (Figure 1). This is the first effort to locate this sequence in Hymenoptera. The presence of a unique chromosomal cluster of U2 snDNA was also observed in true bugs (Bardella et al. 2016) and the grasshoppers Abracris flavolineata and Schistocerca gregaria (Bueno et al. 2013; Camacho et al. 2015), although multiple clusters were also noticed in other species, like grasshoppers (Palacios-Gimenez et al. 2013; Castilho et al. 2017). The use of this probe in other Melipona and Hymenoptera will elucidate the organization and evolution patterns of this sequence, which could be a good tool for understanding chromosome evolution.
The heterochromatin was distributed along the chromosomal arms, except in the terminal regions (Figure 2a), which has been observed in specimens from other colonies and is putatively caused by high heterochromatin additions (Rocha and Pompolo 1998; Rocha et al. 2002; Rocha et al. 2007). According to Rocha et al. (2002), heterochromatin amplification could have a crucial role during chromosomal evolution in bees. The differences in the heterochromatin amounts and distribution are remarkable aspects of Melipona, and based in these differences, Rocha and Pompolo (1998) divided the genus into two groups (see introduction). Species with high heterochromatic contents also display larger genome sizes, such as M. scutellaris, whose genome is 1.08 pg (Tavares et al. 2010). The notion of the amplification of repetitive DNAs, which is associated with large heterochromatic blocks, is reinforced in this work by the analysis of the DOP-PCR and C0t-DNA fraction probe hybridization that labeled the same regions corresponding to heterochromatin (Figure 2b, c).
Mitotic metaphases of Melipona scutellaris. a Heterochromatin location revealed by C-banding. Note large amount of heterochromatin distributed along almost the entire chromosomes and euchromatin restrict to terminal of chromosomes. b Chromosomal mapping through fluorescence in situ hybridization (FISH) using as probe the C0t-DNA fraction of M. scutellatis. c FISH with the DOP-PCR probe obtained from the genomic DNA of M. scutellaris. Note the spread marks along the heterochromatic regions.
In contrast to the mapping with the C0t-DNA and DOP-PCR probes, seven microsatellite repeats revealed enrichment in the euchromatic regions, although faint scattered signals were also observed in the heterochromatin. No remarkable variability in density was observed in the distinct chromosomes (Figure 3). In other insects, euchromatic locations for some microsatellite repeats have been reported, such as in grasshoppers and crickets (Milani and Cabral-de-Mello 2014; Ruiz-Ruano et al. 2015; Palacios-Gimenez and Cabral-de-Mello 2015). In the Hymenoptera model species Nasonia vitripennis, the microsatellite composition, abundance, and density in distinct chromosomes were analyzed using genomic data. The data contrasted with observations in this study based on the density heterogeneity of microsatellites in the five chromosomes from this species. Moreover, considering the heat map for the chromosomal distribution of microsatellites, there are no common regions with higher microsatellite density (Pannebakker et al. 2010) as observed for the distal euchromatic regions in M. scutellaris. It has been documented that the genome size could correlate with microsatellite length and frequency. The presence of several microsatellites contributes to increased genome sizes in many organisms, although it is not a general rule (Hancock 1996, 2002; Butcher et al. 2000; Tóth et al. 2000; Warner and Noor 2000; Comeron 2001). Considering the enrichment for microsatellites in M. scutellaris euchromatin and that the genome increase is related to heterochromatin amplification, we could speculate that the microsatellite motifs mapped here play a minor role in genome size increase in this species. Moreover, the microsatellite mapping indicates the sharing of repetitive DNAs enriched in euchromatin (C-negative regions), similar to what was observed for M. rufiventris heterochromatin (Lopes et al. 2014). Putatively, the repetitive DNAs associated with heterochromatin amplification in M. scutellaris are satDNAs and transposable elements, which are generally isolated in C0t-DNA fraction. However, as C0t-DNA is anonymous, specific sequences should be characterized to ascertain it.
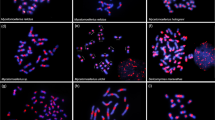
Melipona species with high heterochromatin amounts are placed into two distinct subgenera (Michmelia and Melikerria). Based on phylogenetic data, Lopes et al. (2011) proposed that heterochromatin amplification occurred more than once in Melipona. Our data using dot-blot hybridization shed light on this question. Membrane hybridization using the C0t-DNA fraction of M. scutellaris as a probe against the genomic DNA of some Melipona species (M. rufiventris, M. seminigra, M. bicolor, M. fasciculate, and M. quadrifasciata) only showed positive hybridization to M. rufiventris and M. seminigra, which are species belonging to the same subgenus (Michmelia) (Figure 4). These data support the presence of monophyletic heterochromatin amplification and conservation of sequence similarity among species of Michmelia. These findings also suggest more than one round of heterochromatin amplification, i.e., independent amplification of distinct repetitive DNAs, in distinct subgenera of Melipona or sequence diversification between subgenera. The occurrence of a polyphyletic subgenus (Eomelipona) and of species not assigned to a specific subgenus (Ramirez et al. 2010) is a challenge for a more conclusive hypothesis. The analysis of other species and the better understanding of taxonomy and phylogenetic relationships between Melipona subgenera could shed light on the hypothesis about heterochromatin amplification. Even though the pool of repetitive DNAs is shared between Michmelia species, the specific sequences responsible for heterochromatin amplification should be better investigated, using for example, genome-sequencing data. This is an interesting issue to be investigated in the future and will allow for a more conclusive hypothesis for the causes and the sequences involved in heterochromatin amplification in Melipona. These data together with the chromosomal mapping of repetitive DNAs studied here contribute to the improvement of the hitherto poor knowledge of chromosomal organization of repetitive DNAs among bees, mostly specifically in Melipona species.
Dot-blot hybridization using M. scutellaris C0t-DNA fraction (repetitive DNA enriched fraction) as a probe in species from the Melipona genus groups, group I and group II. a Note the positive signals in M. rufiventris and M. seminigra, which belong to the subgenus Michmelia and have high heterochromatin contents. b Note the absence of signals in species belonging to the other subgenus, including a species with high heterochromatic content, M. (Melikerria) fasciculata. Species with high heterochromatin amount are identified by (+HC) while species with low heterochromatin amount by (−HC).
References
Anjos, A., Rocha, G.C., Paladini, A., Mariguela, T.C., Cabral-de-Mello, D.C. (2016) Karyotypes and repetitive DNA evolution in six species of the genus Mahanarva (Auchenorrhyncha: Cercopidae). Cytogenet. Genome Res. 149, 321–327
Bardella, V.B., Fernandes, J.A.M., Cabral-de-Mello, D.C. (2016) Chromosomal evolutionary dynamics of four multigene families in Coreidae and Pentatomidae (Heteroptera) true bugs. Mol. Genet. Genomic 291, 1919–1925
Brito, R.M., Pompolo, S.G., Magalhães, M.F.M., Barros, E.G., Sakamoto-Hojo, E.T. (2005) Cytogenetic characterization of two Partamona species (Hymenoptera, Apidae, Meliponini) by fluorochrome staining and localization of 18S rDNA clusters by FISH. Cytologia 70 (4), 373–380
Bueno, D., Palacios-Gimenez, O.M., Cabral-de-Mello, D.C. (2013) Chromosomal mapping of repetitive DNAs in the grasshopper Abracris flavolineata reveal possible ancestry of the B chromosome and H3 histone spreading. PLoS One 8 (6), e66532
Butcher, R.D., Hubbard, S.F., Whitfield, W.G. (2000) Microsatellite frequency and size variation in the parthenogenetic parasitic wasp Venturia canescens (Gravenhorst) (Hymenoptera: Ichneumonidae). Insect Mol Biol 9, 375–384
Cabral-de-Mello, D.C., Moura, R.C., Martins, C. (2010) Chromosomal mapping of repetitive DNAs in the beetle Dichotomius geminatus provides the first evidence for an association of 5S rRNA and histone H3 genes in insects, and repetitive DNA similarity between the B chromosome and A complement. Heredity 104, 393–400
Camacho, J.P.M., Ruiz-Ruano, J., Martin-Blázquez, R., Lópes-León, M.D., Cabrero, J., Lorite, P., Cabral-de-Mello, D.C.; Bakkali, M. (2015) A step to the gigantic genome of the desert locust: chromosomes sizes and repeated DNAs. Chromosoma 124 (2), 263–275
Camargo, J.M.F. (2013) Historical biogeography of the Meliponini (Hymenoptera, Apidae, Apinae), in: Vit, P., Pedro, S. R. M., Roubik, D. (Eds.) Pot-Honey: A legacy of stingless bees. Springer-Verlag, New York, pp. 19–34
Castilho, E.R.D., Taffarel, A., Maronna, M.M., Cigiliano, M.M., Palacios-Gimenez, O. M., Cabral-de-Mello, D. C., Martí, D. A. (2017) Phylogeny and chromosomal diversification in the Dichroplus elongates species group (Orthoptera, Melanoplinae). PLoS One 12 (2), e0172352
Charlesworth, B., Snlegowskl, P., Stephan, W. (1994) The evolution dynamics of repetitive DNA in eukaryotes. Nature 371 (6494), 215–220
Comeron, J.M. (2001) What controls the length of noncoding DNA? Curr Opin Genet Dev 11, 652–659
Gokhman, V.E., Anokhin, B.A., Kuznetsova, V.G. (2014) Distribution of 18S rDNA sites and absence of the canonical TTAGG insect telomeric repeat in parasitoid Hymenoptera. Genetica 142 (4), 317–322
Heard, T.A. (1999) The role of stingless bees in crop pollination. Annu. Rev. Entomol. 44, 183–206
Hancock, J.M. (1996) Simple sequences and the expanding genome. Bioessays 18, 421–425
Hancock, J.M. (2002) Genome size and the accumulation of simple sequence repeats: implications of new data from genome sequencing project. Genetica 115, 93–103
Imai, H.T., Taylor, R.W., Crossland, M.W.L., Crozier, R.H. (1988) Modes of spontaneous chromosomal mutation and karyotype evolution in ants with reference to minimun interaction hypotesis. Jpn. J. Genet. 63 (2), 159–185
Imai, H.T., Satta, Y., Takahana, N. (2001) Integrative study on chromosome evolution of mammals, ants and wasps based on the minimum interaction theory. J. Theor. Biol. 210 (4), 475–497
Lopes, D.M., Fernandes, A., Praça-Fontes, M.M., Werneck, H.A., Resende, C., Campos, L.A.O. (2011) Cytogenetics of three Melipona species (Hymenoptera, Apidae, Meliponini). Sociobiology 57 (2)
Lopes, D.M., Fernandes, A., Diniz, D., Sobrinho Scudeler, P.E., Foresti, F., Oliveira Campos, L. A. (2014) Similarity of heterochromatic regions in the stingless bees (Hymenoptera: Meliponini) revealed by chromosome painting. Caryologia 67 (3), 222–226
López-Flores, I., Garrido-Ramos, M.M. (2012) The repetitive DNA content of eukaryotic genomes. Cytogenet. Genome Res. 7, 1–28
Martins, C. (2007) Chromosomes and repetitive DNAs: a contribution to the knowledge of fish genome. In: Pizano, E., Ozouf-Costaz, C., Forest, F., Kapoor, B.G. Fish Cytogenetics, Science Published, Hauppauge, p. 421–453
Menezes, R.S.T., Silva, T.M., Carvallho, A.F., Andrade-Souza, V., Silva, J.G., Costa, M.A. (2013) Numerical and structural chromosome variation in the swarn-founding wasp Metapolybia decorata Gribodo 1896 (Hymenoptera, Vespidae). Genetica 141 (7–9), 273–280
Michener, C.D. (2007) The bees of the world. Johns Hopkins University Press, Baltimore.
Michener, C.D. (2013) The Meliponini, in: Vit, P., Pedro, S.R.M., Roubik, D. (Eds.) Pot-Honey: A legacy of stingless bees. Springer-Verlag, New York, pp. 19–34
Milani, D., Cabral-de-Mello, D.C. (2014) Microsatellite organization in the grasshopper Abracris flavolineata (Orthoptera: Acrididae) revealed by FISH mapping: remarkable spreading in the A and B chromosomes. PLoS One 9 (5), e97956
Nowak, R. (1994) Mining Treasures From ‘Junk DNA’. Science 263, 608–610
Palacios-Gimenez, O.M., Cabral-de-Mello, D.C. (2015) Repetitive DNA chromosomal organization in the cricket Cycloptiloides americanus: a case of the unusual X1X20 sex chromosomes system in Orthoptera. Mol. Genetics Genomics 290, 623–631
Palacios-Gimenez, O.M., Castilho, E.R., Marti, D.A., Cabral-de-Mello, D.C. (2013) Tracking the evolution of sex chromosome systems in Melanoplinae grasshoppers through chromosomal mapping of repetitive DNA sequences. BMC Evol. Biol. 13,167
Paladino, L.C., Papeschi, A., Lanzavecchia, S., Cladera, J., Bressa, M.J. (2013) Cytogenetic charactezation of Diachasmimorpha longicaudata (Hymenoptera: Braconidae), a parasitoid wasp used as a biological control agente. Eur. J. Entomol. 110 (3), 401–409
Pannebakker, B.A., Niehuis, O., Hedley, A., Gadau, J., Shuker, D.M. (2010) The distribution of microsatellites in the Nasonia parasitoid wasp genome. Insect Mol. Biol. 19 (1), 91–98
Pedro, S.R.M. (2014) The Stingless Bee Fauna In Brazil (Hymenoptera:Apidae). Sociobiology 61 (4), 348–354
Pinkel, D., Straume, T., Gray, J.W. (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. U S A 83 (9), 2934–2938
Ramirez, S.R., Nieh, J.C., Quental, T.B., Roubik, D.W., Imperatriz-Fonseca, V. L., Pierce, N. E. (2010) A molecular phylogeny of the stingless bee genus Melipona (Hymenoptera: Apidae). Mol. Phylogenet. Evol. 56 (2), 519–525
Rocha, M.P., Pompolo, S.G. (1998) Karyotypes and heterochromatin variation (C-bands) in Melipona species (HYMENOPTERA, APIDAE, MELIPONINAE). Genet. Mol. Biol. 21 (1)
Rocha, Marla Piumbini. Análises citogenéticas em abelhas do gênero Melipona (Hymenoptera, Meliponini). 2002, 84 f. Dissertação (Doutorado em Biologia Celular e Estrutural) – Curso de Pós-Graduação em Biologia Celular e Estrutural, Universidade Estadual de Campinas, Campinas, 2002.
Rocha, M.P., Pompolo, S.G., Dergam, J.A., Fernandes, A., Campos, L.A.O. (2002) DNA characterization and karyotypic evolution in the bee genus Melipona (Hymenoptera, Meliponini). Hereditas 136, 19–27
Rocha, M.P., Pompolo, S.G., Fernandes, A., Campos, L.A.O. (2007) Melipona: seis décadas de citogenética. J. Biosci. 23, 182–276
Roubik, D.W. (2006) Stingless bee nesting biology. Apidologie 37 (2), 124–143
Ruiz-Ruano, F.J., Cuadrado, A., Montiel, E.E., Camacho, J.P., López-Leon, M.D. (2015) Next generation sequencing and FISH reveal uneven and nonrandom microsatellite distribution in two grasshopper genomes. Chromosoma 124 (2), 221–234
Sambrook, J., Russel, D.W. (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York.
Sumner, A.T. (1972) A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75 (1), 304–306
Tavares, M.G., Carvalho, C.R., Soares, F.A.F. (2010) Genome size variation in Melipona species (Hymenoptera: Apidae) and sub-grouping by their DNA contente. Apidologie 41 (6), 636–642
Tavares, M.G., Lopes, D.M., Campos, L.A.O. (2017) An overview of cytogenetics of the tribe Meliponini (Hymenoptera: Apidae). Genetica 145 (3), 241–258
Telenius, H., Carter, H.P., Bebb, C.E., Nordenskjöld, M., Ponder, P.A. Tunnacliffe, A. (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13 (3), 718–725
Tóth, G., Gaspari, Z., Jurka, J. (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10, 967–981
Warner, R.D., Noor, M.A. (2000) High frequency of microsatellites in Drosophila pseudoobscura. Genes Genet Syst 75, 115–118
Zwich, M.S., Hanson, R.E., Mcknight, T.D. Islam-Faridi, H.M., Stelly, D.M., Wing, R. A., Price, J.H. (1997) A rapid procedure for the isolation of C 0 t-1 DNA from plants. Genome 40 (1), 138–142
Acknowledgements
This study was partially supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (process number 2014/11763-8), Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). DCCM was the recipient of a research productivity fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (process number 304758/2014-0), VBB was a recipient of PNPD/CAPES scholarship and MCAP was recipient of CAPES scholarship.
Author information
Authors and Affiliations
Contributions
DCCM conceived the study and designed the experiments; MCAP and VBB performed the experiments; MCAP, VBB, and DCCM interpreted the data and wrote the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Manuscript editor: Marina Meixner
ADN répétitifs chez Melipona scutellaris (Hymenoptera: Apidae: Meliponidae): distribution chromosomique et test d'amplification multiple de l'hétérochromatine dans le genre
abeilles / FISH/ hétérochromatine / familles multigéniques / ADN répétitifs
Repetitive DNA in Melipona scutellaris (Hymenoptera: Apidae: Meliponidae): Verteilung über die Chromosomen und Test auf multiple Amplifikation von Heterochromatin innerhalb der Gattung
Biene / FISH/ Heterochromatin / Multigenfamilien / repetitive DNA
Rights and permissions
About this article
Cite this article
Piccoli, M.C.A., Bardella, V.B. & Cabral-de-Mello, D.C. Repetitive DNAs in Melipona scutellaris (Hymenoptera: Apidae: Meliponidae): chromosomal distribution and test of multiple heterochromatin amplification in the genus. Apidologie 49, 497–504 (2018). https://doi.org/10.1007/s13592-018-0577-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-018-0577-z