Abstract
During storage, the viability of sperm in a honey bee (Apis mellifera) queen’s spermatheca can be decreased by reactive oxygen species. We hypothesized that the expression of antioxidant genes would increase in queen spermathecae after mating. We measured queen morphometric characteristics and expression levels of seven antioxidant-encoding genes in virgin and mated queen spermathecae. We identified a 12% increase in body weight and a fourfold increase in ovary weight in mated queens. There was a twofold higher expression of catalase, thioredoxin 2, and thioredoxin reductase 1 in the spermathecae of mated vs. virgin queens. Expression of the other antioxidant genes (glutathione S-transferase D1, superoxide dismutase 1, vitellogenin, and glyoxalase domain-containing 4-like (GLOD4L) in spermathecae was not different between mated and virgin queens. In drone semen, expression of antioxidant genes was overall low compared to queens except for GLOD4L, which was equivalent to that in queen spermathecae. Increased expression of antioxidant genes may assist in maintaining sperm viability inside the spermathecae of mated queens.
Similar content being viewed by others
1 Introduction
Reproduction requires direct interactions between sperm and eggs, and between the molecules that comprise the male seminal fluid and glandular and epithelial cells in the female reproductive organs (Heifetz and Rivlin 2010). The reproductive tract of the honey bee (Apis mellifera) queen is divided into upper and lower sections. The upper section contains the ovaries and oviducts, while the lower section contains the sperm storage organ (spermatheca), the accessory glands, and the genitalia. Sperm management (storage, maintenance, and release from storage) and fertilization occurs in the queen’s lower reproductive tract (Snodgrass 1985; Heifetz and Rivlin 2010). Despite the important role that sperm storage plays in reproduction, the molecular mechanisms by which sperms are maintained in a viable state inside the female spermathecae remain unknown.
Virgin honey bee queens mate with 10 to 15 drones in quick succession during one or two mating flights (Tarpy et al. 2004). The queen keeps less than 10% of the semen (Koeniger 1986) and only about 3% of the sperm actively migrate to the spermatheca for long-term storage over the queen’s two- to three-year life span (Baer 2005). This amounts to approximately 4 to 7 million sperm cells in the spermatheca (Harbo 1986; Wilde 1994; Cobey 2007) that the queen uses to fertilize 1500–2000 eggs daily to produce female worker offspring (Winston 1987). Upon mating, the queen’s abdomen and oviduct contract, transporting sperm cells to the spermaduct’s orifice where most cells actively migrate to the spermatheca (Koeniger 1986). However, when live sperm cells are present during fertilization, they can “drag” dead sperm cells along with them to the spermatheca, and thus, live as well as dead sperm cells can be stored in the spermatheca after mating (Collins 2000).
Sperm viability is crucial for reproductive success because it sets the stage for sperm to sperm competition and thus generates selective pressures on male reproductive traits, including flight stamina to achieve copulation, and high sperm viability to maximize storage capacity in competition with other ejaculates (Berg et al. 1997; Collins and Donoghue 1999; Stürup et al. 2013). Factors in seminal fluid have been demonstrated to be crucial for sperm viability in several insect species. In Drosophila melanogaster, the seminal fluid of one male partner improves survival of all sperm collected by a female during mating (Holman 2008). In honey bees, sperm viability can be improved in vitro by incubating sperm with secretions from male accessory glands (components of seminal fluid) or spermathecae (den Boer et al. 2009). In addition, honey bee sperm incubated in vitro with untreated seminal fluid exhibits higher viability than sperm incubated in seminal fluid with heat-denatured proteins (King et al. 2011). Together, these data demonstrate the importance of seminal fluid proteins in maintaining viable sperm for long-term storage.
The metabolic activity of sperm cells during storage appears to be very important for survival. Aerobic metabolism by sperm in the spermathecae creates a dilemma: the sperm needs to produce energy to remain viable during long-term storage, but reactive oxygen-species (ROS) byproducts are produced during this time, which can lower sperm viability (Pardini 1995; Monaghan et al. 2009). Proteins that may support sperm viability have been identified through proteomic analyses of seminal fluid and spermathecal contents collected from honey bee queens before and after mating. Antioxidant proteins are present in higher quantities in the spermathecae of mated queens compared to virgin queens, and they also are present in the seminal fluid of drones (Weirich et al. 2002; Collins et al. 2004; Collins et al. 2006; Baer et al. 2009a; Baer et al. 2009b; den Boer et al. 2009; Gorshkov et al. 2015). These data suggest that an environment capable of protecting against oxidative damage can help maintain sperm viability in mated queens.
Of the 38 antioxidant genes in the honey bee genome (Corona and Robinson 2006), expression of catalase, glutathione-S-transferase D1 (GSTD1), and superoxide dismutase 1 (SOD1) genes has been investigated in the spermathecae of mated and virgin queens (Collins et al. 2004). In that study, levels of catalase and GSTD1 mRNAs were greater in the spermathecae of mated queens compared to virgin queens. In this study, we quantified the relative levels of seven antioxidant-encoding mRNAs in the spermathecae of mated and virgin honey bee queens of Apis mellifera ligustica and A. m. carnica strains, as well as in semen from unmated drones. The selected mRNAs encoded catalase, glyoxalase domain-containing protein 4-like (GLOD4L), GSTD1, SOD1, thioredoxin 2 (TXN2), thioredoxin reductase 1 (TXNRD1), and vitellogenin. All of the seven gene products have roles in antioxidant pathways in honey bees (Collins et al. 2004; Seehuus et al. 2006; Baer et al. 2009a). Our hypothesis was that the expression of specific antioxidant-encoding genes is upregulated in spermathecae after mating to protect sperm from damage by ROS during storage. We identified upregulation of catalase, TXN2, and TXNRD1 genes in the spermathecae of mated queens compared to those of virgin queens. Since drone semen had low levels of those mRNAs, it appears that the spermatheca upregulates expression of those three antioxidant-encoding genes after mating.
2 Materials and methods
2.1 Source of bees
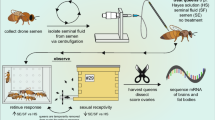
We obtained mated and virgin honey bee queens from a commercial queen producer located in the central valley of California (Olivarez Honey Bees, Inc., Orlando, CA). Roughly half of the queens were of “Italian” stock (presumably A. m. ligustica) and half were of “Carniolan” stock (presumably A. m. carnica). All of the queens of each stock were obtained from the same source colony and thus were sisters to each other. The individually labeled and caged queens were shipped to our laboratory facility surrounded by worker attendants and food. They were kept with attendants at room temperature until they were used for morphometric measurements. Sexually mature drones were collected from a designated hive located at the Janice and John G. Thomas Honey Bee Facility of Texas A&M University’s RELLIS Campus in Bryan, TX. They were placed in a cage for less than 24 h until semen was collected.
2.2 Morphological measures
Each queen was immobilized by placing her in a -20 °C freezer for 3 to 5 min. Once immobilized, her fresh weight was measured to the nearest 0.1 mg on a digital scale. Then, her head and thorax widths were measured three times to within 0.1 mm using digital calipers, and the three-measurement averages were calculated. All queens were sacrificed by decapitation on a bed of dry ice to keep all tissues suitable for RNA extraction. We then dissected each queen’s ovaries and spermatheca from the abdomen. The wet weight of the two ovaries was measured to the nearest 0.1 mg on a digital scale. The tracheal net was removed from the spermatheca with forceps and then the spermatheca was pressed against the wall of an Eppendorf tube to release the contents therein. A small amount of semen from four mated Italian and four mated Carniolan queens was collected and sampled immediately for sperm viability analysis. The rest of the spermathecal samples from virgin and mated queens were frozen individually at -80 °C until they were used for RNA extraction (see below). Likewise, drones were sacrificed by decapitation. Semen was collected from each drone by everting the endophallus and by using a micropipette to harvest the semen on top of the mucus, as previously described (Cobey 2007). The drone semen samples were frozen individually at -80 °C until they were used for RNA extraction (see below).
2.3 Sperm counts and viability
We used a staining protocol following the general guidelines outlined by Collins and Donoghue (1999) to quantify sperm number and viability in the semen collected from the spermathecae of mated queens. A LIVE/DEAD Sperm Viability Kit from Molecular Probes (Eugene, OR) was used with the following modifications: Briefly, immediately after dissection, the queen’s spermatheca was placed in a microcentrifuge tube with 200 μL of 10 mM HEPES, 150 mM NaCl, and 10% BSA, pH 7.4. The spermatheca was squeezed with forceps to break it open and release the sperm. The spermathecal tissue was allowed to settle before we harvested 100 μL from the top of the solution for the sperm counting and viability assay. To perform the sperm viability assay, we added 6 μL of 1 mM SYBR 14 in DMSO and 6 μL of 2.4 mM propidium iodide in water to the 100-μL solution containing sperm and incubated it for 8 min in the dark. Brightfield cell counting was used to estimate sperm cell concentration, while dual fluorescent filters were used to measure sperm viability by estimating the proportion of live sperm in the total cell count for each sample. Live sperm fluoresce at green wavelengths (520 to 570 nm), while dead sperm fluoresce at red wavelengths (approximately 650 nm; Collins and Donoghue 1999). The stained sperm solution (20 μL) was immediately loaded onto a slide and allowed to settle for 5 min before conducting the automated cell-counting analysis using a Vision CBA Analysis System® (Nexcelom Bioscience; Lawrence, MA). The average number of live and dead sperm cells was counted twice for each sample, and the average was corrected for the dilution factor to estimate the total number of sperm cells.
2.4 RNA purification and production of complementary DNA (cDNA)
Spermathecae were individually placed in a 1.5-mL microtubes with 50 μL PureZOL™ RNA isolation reagent (BIO-RAD). The same RNA isolation procedure was done for semen collected from drones. The tissues in PureZOL were ground with a VWR® pellet mixer (catalog number 47747-307, VWR®, Radnor, PA). We followed the standard RNA-extraction protocol for PureZOL™. The precipitation step was done overnight at -20 °C after adding 1 μL of 20 μg/μL RNAse-free glycogen. RNA was pelleted during 10 min of centrifugation at 12,000×g in a 4 °C microcentrifuge. RNA pellets were washed in 75% ethanol, air dried for 10 min, and resuspended in 25 μL of nuclease-free water. RNA concentrations were measured in a Qubit®2.0 fluorometer with a Qubit® RNA HS Assay Kit (Life Technologies Corporation, Grand Island, NY). Fifty nanograms of each RNA sample was processed to remove genomic DNA with an iScript™ gDNA Clear cDNA Synthesis Kit (BIO-RAD)®. Then, the kit, employing both oligo dT and random primers, was used to produce 20 μL of cDNA per sample.
2.5 Quantitative PCR analysis
The cDNA samples were diluted in a 1:4 ratio with nuclease-free water, and 1 μL of the diluted sample was used to perform replicate 10-μL quantitative-PCR (qPCR) reactions. The primers utilized to amplify each gene product (Table I) were synthesized by Integrated DNA Technologies (Coralville, IA). All newly designed primers were made to amplify across a large intron to add specificity for amplification of cDNA and not genomic DNA. The qPCR reactions were performed with the SsoAdvanced™ Universal SYBR® Green Supermix kit, which uses a Sso7d fusion polymerase enzyme (BIO-RAD®). The qPCR was performed in a CFX96™ Real-time system (BIO-RAD®). The cycling program was 95 °C for 30 s followed by 39 cycles at 95 °C for 5 s, 61 °C for 10 s, followed by a melting curve from 65 to 95 °C in 0.5 °C increments. All of the amplicons melted at their individual predicted temperatures. Gene expression was normalized to the expression of the normalizer genes RPL8 and RPS5. Normalized values were calculated utilizing the CFX Manager™ Software and reported as “2−ΔΔCt” values. The coefficient of variation (CV%) and average expression stability (M value) of the tested genes were CV = 0.1702 and M = 0.4826.
2.6 Statistical analysis
We used one-way ANOVA tests to determine whether there were differences in the morphometric measurements of Carniolan and Italian queens. We also performed one-way ANOVA tests to determine if there were statistical differences in gene expression values between spermathecae of virgin and mated queens and drone semen. In cases where we found statistical differences between queens and drones, we conducted pairwise Student’s t tests to discern differences between two tissue types. All tests were performed using the statistical software JMP® 12.0 (SAS Inc., Cary, NC). We set the level of statistical significance at α = 0.05 for all tests and report all descriptive statistics as means ± standard errors of the mean (S.E.M.).
3 Results
3.1 Morphometric, gene-expression, and sperm measurements in spermathecae were not different between Italian and Carniolan queens
We found no statistically significant differences between virgin Carniolan and virgin Italian queens for any of the morphometric measurements or any of the gene expression values for the seven genes measured (Table II). However, there was a trend (P = 0.07) for a fourfold higher level of GSTD1 gene expression in the spermathecae of virgin Italian queens compared to virgin Carniolan queens. Likewise, we did not find any statistical significant differences for any of the morphometric characters or normalized gene expression values measured from mated Carniolan and mated Italian queens (Table II). Furthermore, we did not find statistical differences in spermathecal sperm counts or sperm viability between Carniolan and Italian mated queens (Table II). Based on these results, we grouped all virgin-queen and mated-queen data into single categories for further statistical analysis, regardless of the genetic stock of the queens.
3.2 Morphometric differences between virgin and mated queens
For most morphometric measurements, we collected data from 10 virgin and 18 mated queens, unless otherwise noted in the tables. Not surprisingly, the wet weight of mated queens was on average 12% greater than that of virgin queens (Table III). We found no differences in head or thorax width between virgin and mated queens (Table III). Also, not surprisingly, the wet weight of the ovaries showed the greatest change in mated queens, as it was more than five times higher that of virgin queens. On average for all mated queens analyzed, the spermathecal sperm counts were (6.83 ± 0.44) × 106 sperm cells and the sperm viability was 94.44 ± 1.47% (Table II).
3.3 Identified differences in expression levels of seven antioxidant genes in the spermathecae of mated and virgin queens and drone semen
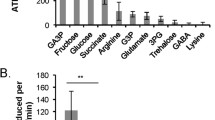
Figure 1 reports normalized expression (2−ΔΔCt) values for the seven antioxidant-encoding genes explored in the spermathecae of mated and virgin queens and in drone semen (mean ± S.E.M.). We identified statistically significant differences across all three sample types for the expression of catalase (F 2,27 = 6.55, P = 0.005), SOD1 (F 2,28 = 10.01, P value = 0.0005), TXN2 (F 2,28 = 14.95, P < 0.0001), and TXNRD1 (F 2,28 = 7.67, P = 0.002) genes. Conversely, we did not find any statistical differences across all three sample types for the expression of GLOD4L (F 2,28 = 0.10, P = 0.90), GSTD1 (F 2,24 = 2.80, P = 0.08), and vitellogenin (F 2,23 = 0.36, P = 0.70) genes.
Normalized expression of seven antioxidant genes in the spermathecae of mated and virgin queens and the semen of sexually mature drones (panels a–g). Gene expression was normalized to the expression of the normalizer genes RPL8 and RPS5 and reported as the mean ± standard error of the mean (S.E.M.). Statistically significant pairwise differences between two tissue groups are represented by connecting lines and asterisks above the bars. * Values of P ≤ 0.05. ** Values of P ≤ 0.001. *** Values of P ≤ 0.0001. See Section 2 for details.
For those genes that revealed significant differences across all three samples, further pairwise comparisons between tissue types revealed that expression of the catalase gene was almost two times greater in the spermathecae of mated queens compared to that of virgin queens (t = − 2.31, P = 0.03) and 15 times greater than that in drone semen (t = 3.36, P = 0.002; Figure 1a). Expression of the TXN2 gene was almost two times greater in the spermathecae of mated queens than in those of virgin queens (t = − 2.65, P = 0.01) and five times greater than that in drone semen (t = 5.38, P < 0.0001; Figure 1d). Moreover, expression of the TXN2 gene was three times greater in spermathecae of virgin queens compared to drone semen (t = 2.18, P = 0.03). The expression levels of the functionally related TXNRD1 gene in the spermathecae of mated queens were almost three times greater than in the spermathecae of virgin queens (t = − 2.90, P = 0.007; Figure 1d) as well as in the semen of drones (t = 3.42, P = 0.002; Figure 1e). Likewise, levels of SOD1 mRNA were four times greater in the spermathecae of mated queens compared to drone semen (t = 3.97, P = 0.0004) and almost four times greater in the spermathecae of virgin queens compared to drone semen (t = 3.92, P = 0.0005; Figure 1f).
Interestingly, although the overall expression of the GSTD1 gene was not statistically significant across all three tissue types (P = 0.08), its expression was 7.5 times greater in the spermathecae of virgin queens compared to drone semen (t = 2.31, P = 0.03; Figure 1c). We did not find significant pairwise differences in the expression of GLOD4L (Figure 1b) or vitellogenin (Figure 1g) genes in the tissues analyzed. As expected, there was negligible expression of the vitellogenin gene in drone semen compared to that in the spermathecae of virgin and mated queens. The variation in vitellogenin gene expression between individual spermathecae precluded identification of a difference between spermathecal values and those of semen. The concentrations of catalase, TXN2, and TXNRD1 mRNAs were three to 14 times lower in drone semen than in the spermathecae of mated queens (Figure 1). Together, the data indicate that honey bee spermathecae upregulate expression of at least four antioxidant-encoding genes after mating.
4 Discussion
Apis mellifera ligustica (“Italian”) and A. m. carnica (“Carniolan”) are honey bee subspecies that come from regions near the Mediterranean Sea. Despite their close geographical precedence, they differ phenotypically and in behavior. For example, Italian bees, the dominant subspecies introduced into North America and Europe, are yellowish-brown and exhibit low swarming tendencies. In contrast, the Carniolan bees are darker in color, better house cleaners, less aggressive, and more resistant to disease (Caron and Connor 2013). Even though there are genetic differences between these two subspecies, we found no differences between Italian and Carniolan queens morphometrically or in spermathecal expression of antioxidant-encoding genes. Not surprisingly, in both subspecies, ovary and wet body weights were greater in mated compared to virgin queens, which is explained by the fact that ovary development is triggered by mating (Winston 1987).
We discovered higher levels of expression of the catalase, TXN2, and TXNRD1 genes in spermathecae of mated queens compared to those of virgin queens. Likewise, Weirich et al. (2002) found catalase, GSTD1, and SOD1 to be catalytically active enzymes in honey bee spermathecae. They also reported that activities of catalase and GSTD1 enzymes were elevated in the spermathecae of mated queens compared to virgin queens. In contrast, levels of SOD1 activity were not different between spermathecae of mated and virgin queens. The increase of catalase mRNA levels in the spermathecae after mating has been described by Collins et al. (2004). However, that study also identified an increase in GSTD1 mRNA levels at 21 days post-mating which we did not find, possibly due to differences in the protocols. The 2004 study, which used Italian bees from a Claxton, GA apiary, only found increased levels of GSTD1 mRNA in the queen spermathecae at 21 days post-mating and not at a later time point, so perhaps the effect is transient. Furthermore, our cDNA synthesis utilized both oligo(dT) and random primers while Collins et al. (2004) utilized only oligo(dT) primers. As a consequence of the latter, the reverse transcriptase would have to synthesize 676 bases of GSTD1 cDNA (GenBank accession NM_001178028.1) through the entire 3′ untranslated region and 84% of the coding sequence to produce the amplicon in quantitative PCR. Moreover, our results showed that drone semen contained low levels of all the antioxidant-encoding transcripts we analyzed except GLOD4L mRNA relative to those in queen spermathecae. Therefore, it appears that spermathecae indeed upregulate expression of catalase, TXN2, and TXNRD1 genes after mating.
The expression of antioxidant-encoding genes in reproductive tracts is widely conserved across species from insects, to birds, to mammals (Aitken and Fisher 1994; Bilodeau et al. 2002; Collins et al. 2004; Lapointe and Bilodeau 2003; Seehuus et al. 2006; Tavilani et al. 2008; Heifetz and Rivlin 2010; Malta et al. 2014; Du et al. 2015). Thioredoxins, including TXN2 and TXNRD1, are a family of antioxidant proteins that have been largely conserved from bacteria to mammals (Meyer et al. 2009). There are multiple antioxidant enzymes in the thioredoxin family, which can specifically interact with a target protein and reduce a disulfide bond. In D. melanogaster, the related gene-encoding gluthathione reductase is lacking and thioredoxin-related gene products are the predominant enzymes with antioxidant activities (Kanzok et al. 2001; Koo et al. 2016). For instance, TXN2 mutant fruit flies have greater oxidative stress and shorter life spans (Svensson and Larsson 2007). In mammalian systems, expression of thioredoxin-associated genes appears to be crucial for spermatogenesis and other developmental processes (Meyer et al. 2009). TXNRD1 has two redox-active sites compared to one in glutathione reductase (Mustacich and Powis 2000). Because of this, TXNRD1 has a much greater number of substrates than glutathione reductase. TXNRD1 functions not only in oxidant injury prevention, but also in cell growth, DNA synthesis, and regulation of gene expression. TXN2 is in the mitochondria, which have recently been recognized as regulators of the concentrations of reactive oxygen species (Munro and Treberg 2017).
The honey bee genome has seven genes related to the thioredoxin family (Corona and Robinson 2006). The TXN2 and TXNRD1 proteins were found to be present in the spermathecal fluid of mated queens (Baer et al. 2009a). Our results of greater TXN2, TXNRD1, and catalase expression in the spermathecae of mated compared to virgin queens suggest that the genes may be transcriptionally induced after mating. Factors leading to enhanced expression of these genes may be from sperm or seminal fluid or from physiological alterations of the queen as she initiates egg production. In future studies, it will be exciting to define the molecular mechanism responsible for the upregulation of antioxidant genes in queen spermathecae.
Reproduction is critically important to fitness, as failure to reproduce will compromise species survival. In honey bees, reproduction of the colony depends solely on the reproductive capacity of a single queen, the quality of her drone partners, and the viability of the sperm she stores after mating. Consequently, workers have developed mechanisms to recognize a decrease in the queen’s ability to lay fertilized eggs and to rapidly begin a queen replacement process (i.e., “supersedure”) if the queen fails in her egg-laying duties. Creation of a replacement queen is energetically costly and, if resources are scarce, it may cause a colony to collapse (Rangel et al. 2012). Therefore, it is advantageous for the colony to have long-lived queens with viable sperm in the spermatheca. During storage, sperm metabolism produces ROS, which can impair sperm viability. The expression of antioxidant genes in the spermatheca of mated queens is likely needed to protect sperm from ROS during their long-term storage. Interestingly, Verma and Shuel (1973) discovered that stored semen showed only 40% of the oxygen uptake of freshly ejaculated semen, thus exhibiting significantly lower rates of oxygen consumption and conservation of energy in semen stored in the spermatheca. Therefore, the high density of sperm in the spermatheca, as well as the low metabolic activity that it undergoes during storage, may also play important roles in the queen’s successful long-term storage of sperm. The honey bee queen’s spermatheca presents itself as an example of how evolution can meticulously craft a reproductive organ to overcome strong physiological and environmental constraints to maintain its vigor over time.
References
Aitken, J., Fisher, H. (1994) Reactive oxygen species generation and human spermatozoa: the balance of benefit and risk. Bioessays 16, 259–67
Baer, B. (2005) Sexual selection in Apis bees. Apidologie 36, 187–200
Baer, B., Eubel, H., Taylor, N.L., O’Toole, N., Millar, A.H. (2009a) Insights into female sperm storage from the spermathecal fluid proteome of the honeybee Apis mellifera. Genome Biol, 10: R67
Baer, B., Heazlewood, J.L., Taylor, N.L., Eubel, H., Millar, A.H. (2009b) The seminal fluid proteome of the honeybee Apis mellifera. Proteomics 9, 2085–2097
Berg, S., Koeniger, N., Koeniger, G., Fuchs, S. (1997) Body size and reproductive success of drones (Apis mellifera L). Apidologie 28, 449–460
Bilodeau, J.F., Blanchette, S., Cormier, N., Sirard, M.A. (2002) Reactive oxygen species-mediated loss of bovine sperm motility in egg yolk Tris extender: protection by pyruvate, metal chelators and bovine liver or oviductal fluid catalase. Theriogenology 57, 1105–22
Caron, D.M., Connor, L.J. (2013) Honey Bee Biology and Beekeeping, Revised Edition. Wicwas Press, Kalamazoo, MI
Cobey, S.W. (2007) Comparison studies of instrumentally inseminated and naturally mated honey bee queens and factors affecting their performance. Apidologie 38, 390–410.
Collins, A.M. (2000) Relationship between semen quality and performance of instrumentally inseminated honey bee queens. Apidologie 31, 421–429
Collins, A.M., Donoghue, A.M. (1999) Viability assessment of honey bee, Apis mellifera, sperm using dual fluorescent staining. Theriogenology 51, 1513–1523
Collins, A.M., Williams, V., Evans, J.D. (2004) Sperm storage and antioxidative enzyme expression in the honey bee, Apis mellifera. Insect Mol. Biol. 13, 141–6
Collins, A.M., Caperna, T.J., Williams, V., Garrett, W.M., Evans, J.D. (2006) Proteomic analyses of male contributions to honey bee sperm storage and mating. Insect Mol. Biol. 15, 541–549
Corona, M., Robinson, G.E. (2006) Genes of the antioxidant system of the honey bee: annotation and phylogeny. Insect Mol. Biol. 15, 687–701
den Boer, S.P.A., Boomsma, J.J., Baer, B. (2009) Honey bee males and queens use glandular secretions to enhance sperm viability before and after storage. J. Insect Physiol. 55, 538–543
Du, J., Hincke, M.T., Rose-Martel, M., Hennequet-Antier, C., Brionne, A., Cogburn, L.A., Nys, Y., Gauron, J. (2015) Identifying specific proteins involved in eggshell membrane formation using gene expression analysis and bioinformatics. BMC Genomics 16: 792
Gorshkov, V., Blenau, W., Koeniger, G., Römpp, A., Vilcinskas, A., Spengler, B. (2015) Protein and peptide composition of male accessory glands of Apis mellifera drones investigated by mass spectrometry. PLoS ONE 10:e0125068
Harbo, J.R. (1986) Propagation and instrumental insemination, In: Rinderer T.E. (Ed.), Bee Breeding and Genetics. Academic Press, Inc., Orlando, FL. pp. 361–389
Heifetz, Y., Rivlin, P.K. (2010) Beyond the mouse model: Using Drosophila as a model for sperm interaction with the female reproductive tract. Theriogenology 73, 723–739
Holman, L. (2008) Drosophila melanogaster seminal fluid can protect the sperm of other males. Funct. Ecol. 23, 180–186
Kanzok, S.M., Fechner, A., Bauer, H., Ulschmid, J.K., Muller, H.M., Botella-Munoz, J.Schneuwly, S., Schrmer, R., Becker, K. (2001) substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science 291, 643–646
King, M., Eubel, H., Millar, A.H., Baer, B. (2011) Proteins within the seminal fluid are crucial to keep sperm viable in the honeybee Apis mellifera. J. Insect Physiol. 57, 409–414
Koeniger, G. (1986) Reproduction and mating behavior, in: Rinderer T.E. (Ed.), Bee Breeding and Genetics. Academic Press, Inc., Orlando, FL, pp. 235–252
Koo, H.-N., Lee, S.G., Yun, S.-H., Kim, H.K., Choi, Y.S., Kim G.-H. (2016) Comparative analyses of Cu-Zn superoxide dismutase (SOD1) and thioredoxin reductase (TrxR) at the mRNA level between Apis mellifera L. and Apis cerana F. (Hymenoptera: Apidae) under stress conditions. J. Insect Sci. 16:1–6
Lapointe, J., Bilodeau, J.F. (2003) Antioxidant defenses are modulated in the cow oviduct during the estrous cycle. Biol. Reprod. 68, 1157–64
Malta, J., Martins, G.F., Marques, A.E., Games, P.D., Zanuncio, J.C., Baracat-Pereira, M.C., Salomão, T.M.F. (2014) Insights into the proteome of the spermatheca of the leaf-cutting ant Atta sexdens rubropilosa (Hymenoptera: Formicidae). Fla. Entomol. 97, 1856–1861
Meyer, Y., Buchanan, B.B., Vignols, F., Reichheld, J.-P. (2009) Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 43, 335–67
Monaghan, P., Metcalfe, N.B., Torres, R. (2009) Oxidative stress as a mediator of life history trade-offs: mechanisms, measurements and interpretation. Ecol. Lett. 12, 75–92
Munro, D., Treberg, J.R. (2017) A radical shift in perspective: mitochondria as regulators of reactive oxygen species. J Exper. Biol. 220, 1170–1180
Mustacich, D., Powis, G. (2000) Thioredoxin reductase. Biochem. J. 346, 1–8
Pardini, R.S. (1995) Toxicity of oxygen from naturally occurring redox-active pro-oxidants. Arch. Insect Biochem. 29, 101–118
Rangel, J., Keller, J. J., Tarpy, D. R. (2012) The effects of honey bee (Apis mellifera L.) queen reproductive potential on colony growth. Insectes Soc. 60, 65–73
Seehuus, S.C., Norberg, K., Gimsa, U., Krekling, T., Amdam, G.V. (2006) Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 103, 962–7
Snodgrass, R.E. (1985) Anatomy of the Honey Bee. Cornell University Press, Ithaca, NY
Stürup, M., Baer-Imhoof, B., Nash, D.R., Boomsma, J.J., Baer, B. (2013) When every sperm counts: factors affecting male fertility in the honeybee Apis mellifera. Behav. Ecol. 24, 1192–1198
Svensson, M.J., Larsson, J. (2007) Thioredoxin-2 affects lifespan and oxidative stress in Drosophila. Hereditas 144, 25–32
Tarpy, D.R., Nielsen, R., Nielsen, D.I. (2004) A scientific note on the revised estimates of effective paternity frequency in Apis. Insectes Soc. 51, 203–204
Tavilani, H., Goodarzi, M.T., Vaisi-raygani, A., Salimi, S., Hassanzadeh, T. (2008) Activity of antioxidant enzymes in seminal plasma and their relationship with lipid peroxidation of spermatozoa. Int. Braz. J. Urol. 34, 485–91
Verma, L.R., Shuel, R.W. (1973) Respiratory metabolism of the semen of the honey-bee, Apis mellifera. J. Insect Physiol. 19, 97–103
Weirich, G.F., Collins, A.M., Williams, V.P. (2002) Antioxidant enzymes in the honey bee, Apis mellifera. Apidologie 33, 3–14
Wilde, J. (1994) Comparison of the development and productivity of bee colonies with naturally and instrumentally inseminated queens kept in different conditions before and after the insemination. Zootechnica 39, 135–152
Winston, M.L. (1987) The Biology of the Honey Bee. Harvard University Press, Cambridge, MA
Acknowledgements
We would like to thank Tammy Olivarez for donating the honey bee queens used in our study. We thank ET Ash and Adrian Fisher II for their assistance in collecting drone samples at the Janice and John G. Thomas Honey Bee Facility at Texas A&M University.
Author information
Authors and Affiliations
Contributions
ANG, NI and JR conceived this research, designed experiments and interpreted the data; ANG performed experiments; ANG, NI and JR performed the analysis, wrote the paper and participated in the revisions of it. All authors read and approved the final manuscript.
Funding
This study was funded in part by a USDA-NIFA grant to JR and NI (award number 2015-67013-23170) and the Texas AgriLife Research Hatch Project number TEX09557.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no potential conflict of interest in relation to the study in this paper.
Additional information
Manuscript editor: Peter Rosenkranz
Régulation à la hausse de gènes antioxydants dans les spermathèques de reines d’abeilles ( Apis mellifera ) après accouplement
gène d’enzymes antioxydantes / Apidae / mâle / abeille / spermathèque / reine / sperme
Upregulation von antioxidativen Genen in der Spermatheka von Bienenköniginnen ( Apis mellifera ) nach der Paarung
Antioxidative Enzymgene / Apis mellifera / Drohnen / Bienenkönigin / Spermatheka / Spermien
Rights and permissions
About this article
Cite this article
Gonzalez, A.N., Ing, N. & Rangel, J. Upregulation of antioxidant genes in the spermathecae of honey bee (Apis mellifera) queens after mating. Apidologie 49, 224–234 (2018). https://doi.org/10.1007/s13592-017-0546-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-017-0546-y