Abstract
Honey bees that consume pollen with sublethal levels of the fungicide Pristine® can have reduced pollen digestion, lower ATP synthesis, and in many ways resemble malnourished bees. Reduced nutrient acquisition in bees exposed to Pristine® might be due to the effects of this fungicide on the composition of gut microbial communities. Two experiments were conducted in two separate years to test for the effects of Pristine® on the composition and diversity of bacteria in nurse bee midguts. In the low-dose experiment, bees fed the fungicide had a reduced relative abundance of Gilliamella sp. and the number of bacterial taxa within each individual bee was lower. In the high-dose experiment, the fungicide treatment led to increased relative abundance of Lactobacillus sp. Firm 4 and Firm 5 relative to the control. Presence of the fungicide did not impact the distribution of bacteria among individuals on either the low-or high-dose experiments. Considerable differences in gut microbial communities existed between the two experiments perhaps due to environmental and dietary factors. The effects of Pristine® on the gut microbiota were inconsistent between experiments indicating that exposure rate and environmental conditions can influence the effects of this fungicide on gut microbial communities.
Similar content being viewed by others
1 Introduction
The effects that pesticides might have on non-target organisms can be broad and difficult to anticipate. One reason for this is that most pesticides applied to reduce crop losses from insects and diseases target physiological systems that are shared among the target pest as well as many non-target species. Honey bees are a non-target organism that frequently encounters pesticides especially when colonies are located in and around agroecosystems (Mullin et al. 2010). Exposure to sublethal levels of pesticides might be contributing to colony losses (Dively et al. 2015) that can average 30 % or more each year (Lee et al. 2015b). Preventing colony losses from encounters with pesticides requires more judicious and careful use of these chemicals, and a greater understanding of the harm they might pose.
Though insecticides, especially neonicotinoids, often are implicated in pollinator losses (e.g., Blacquiere et al. (2012) and Gill et al. (2012)), nectar and pollen are more likely to be contaminated with fungicides. These compounds are applied to plants that are in bloom because fungicides are considered safe to bees with regard to acute toxicity (Legard et al. 2001; Yoshimura et al. 2004). Foraging bees can collect resources contaminated with fungicides applied during bloom. The frequency with which bees encounter fungicides is evident in the pervasive presence of these compounds in pollen stores and wax comb. Only compounds used to control Varroa mites are detected more often (Mullin et al. 2010; Pettis et al. 2013).
Recently, the fungicide Pristine® was reported to have sublethal effects on honey bees when ingested at field-relevant concentrations in pollen. Bees fed pollen with Pristine® had reduced protein digestion, higher virus titers, and lower ATP levels than those fed untreated pollen (Campbell et al. 2016; DeGrandi-Hoffman et al. 2015). Pristine® is composed of two fungicides, boscalid and pyraclostrobin. Both compounds have been found in pollen samples from broad surveys of colonies (Mullin et al. 2010, Simon-Delso et al. 2014). Foraging bees can be exposed to Pristine® because it is sprayed during bloom to prevent crop loss from fungal diseases such as brown rot (Janousek and Gubler 2010). In almonds and cherries, applications of this fungicide are recommended during full bloom to reduce blossom rot (http://www.agproducts.basf.us/products/research-library/pristine-fungicide-on-almonds-technicalinformation-bulletin.pdf, last accessed 17 August 2015). Pristine® prevents fungal growth by inhibiting enzymes in the electron transport chain and preventing ATP synthesis (Anke 1995; Kuhn 1984).
ATP production in mitochondria occurs in all eukaryotes, so it was not surprising that bees fed pollen with Pristine® had reduced ATP levels (Campbell et al. 2016; DeGrandi-Hoffman et al. 2015). The effects of Pristine® on digestion and immune response may be explained by the direct effects of the fungicide on ATP synthesis. Many proteases require ATP to function (Gottesman and Maurizi 1992) so the activity of these enzymes could be compromised when bees ingest this fungicide. Fungicides may also impact cellular immunity in cases where mitochondria modulate cell death and innate immune signaling (Arnoult et al. 2009). An alternative explanation for the sublethal effects of Pristine® on protein digestion and immunity might be that the fungicide altered the composition and diversity of gut microbial communities that may contribute to pollen digestion and pathogen defense (Babendreier et al. 2007; Engel et al. 2012; Koch and Schmid-Hempel 2011; Yang et al. 2011). Though bacteria do not have mitochondria, electron transport occurs in the cell membrane and uses ubiquinone and cytochrome b as electron carriers. Both of these electron carriers can be inhibited by the components of Pristine® and might be responsible for the broad spectrum bactericidal properties associated with strobilurins (Bartlett et al. 2002).
Adult honey bee gut bacterial communities are dominated by seven bacterial lineages and eight species (Anderson et al. 2013; Corby-Harris et al. 2014a; Martinson et al. 2011; Moran et al. 2012; Sabree et al. 2012). The bacteria in these communities, particularly members of the Gilliamella, Frischella, Lactobacillus sp. Firms 4 and 5, and Bifidobacterium genera (Engel et al. 2012; Lee et al. 2015a), occur primarily in the hindgut and are enriched for genes related to carbohydrate digestion, including the pollen exine, which encloses a protein-rich interior, and those involved in nectar digestion and energy acquisition. These clades are also some of the most abundant and species rich in the honey bee gut community. If gut microbes facilitate pollen digestion and are negatively impacted by fungicides, pollen digestion could be reduced as a function of decreased gut microbial species richness and diversity on a coarse scale, or by reduced abundance of any of these genera. Though honey bees commonly encounter fungicides in the pollination environment, whether fungicides such as Pristine® impact honey bee gut bacterial communities is an open question. If Pristine® affects the composition of these communities, this could at least partly explain the previously reported effects of this fungicide on protein digestion and virus titers.
The composition of gut microbial communities in honey bees may appear relatively simple, but there can be extensive species- and strain-level diversity. This diversity can change with age (Anderson et al. 2015), season (Hroncova et al. 2015; Ludvigsen et al. 2015), and nutrition (Ludvigsen et al. 2015; Saraiva et al. 2015). In this study, we used high-throughput sequencing to examine the effects of feeding pollen with added Pristine® on the composition and diversity of microbial communities in the gut of nurse bees (i.e., worker bees that care for brood). Our goal was to determine whether taxa with genomes enriched for carbohydrate degradation genes and the abundance of taxa within samples (α-diversity) were impacted by Pristine®. We also tested whether the distribution of bacterial taxa among nurses (β-diversity) was impacted by Pristine®. Because of their potential broad non-target effects, Pristine® and other fungicides should be investigated more comprehensively to determine whether they are safe for use when pollinators are present.
2 Materials and methods
The study was conducted at the Carl Hayden Bee Research Center, Tucson AZ, USA. European honey bees (Apis mellifera ligustica) were used in all experiments. Colonies were started from package bees with a laying queen purchased from Koehnen and Sons Inc. (Glenn, CA).
Preparing pollen for feeding to colonies
The study was conducted using two separate experiments. The first experiment was conducted from September to October of 2013 using a low dose of Pristine®. A second experiment using the higher dose was conducted from August to September of 2014. Pollen fed to the bees in each experiment was sourced from hives placed in the Sonoran Desert in the spring and equipped with pollen traps to capture incoming pollen. We fed colonies pollen that was collected in the year when the experiment was conducted.
Pollen collected in the pollen traps was ground to a fine powder with a coffee grinder (Mr. Coffee model 1DS77, Sunbeam, Boca Raton, FL, USA). To treat the pollen with fungicide, a 350-g portion was spread evenly on a 0.26-m2 tray. Pristine® was sprayed at two concentrations on to the pollen using techniques previously described in DeGrandi-Hoffman et al. (2013). The target doses are within the high and low ranges of Pristine® detected in corbicular loads from foragers (Simon-Delso et al. 2014; DeGrandi-Hoffman et al. 2015). The control pollen was sprayed with distilled water. The pollen was stored at −20 °C until fed to the bees.
Pesticide analysis
Samples of the pollen fed to colonies were analyzed by the USDA-AMS-National Science Laboratory (NSL) in Gastonia, NC, USA, using an official pesticide extraction method (AOAC 2007.01, also known as the QuEChERS method) and analyzed by gas chromatography and liquid chromatography coupled with mass spectrometry detection (GC/MS, GC/MS/MS, LC/MS/MS) to determine concentrations of boscalid and pyraclostrobin. Quantification of pesticide residues was performed using external calibration standards prepared from certified standard reference material. The NSL is ISO 17025 accredited to perform pesticide residue analysis. The low dose of Pristine® had 1990 ppb (1067 ppb boscalid and 920 ppb of pyraclostrobin—low dose) and the high dose 11,410 ppb (7460 ppb boscalid and 3950 pyraclostrobin—high dose). We did not detect boscalid or pyraclostrobin in control pollen in either experiment.
Establishing honey bee colonies
Ten hives were assembled using 3000–4000 bees from the colonies established from package bees. The bees were shaken into a 5-frame nucleus box containing empty drawn comb. A commercially produced and mated European queen (Pendall Apiaries, Stonyford, CA) was introduced into each hive. A sugar feeder containing a 30 % sucrose solution was placed on top of each colony. The hives were placed in an enclosed flight area (EFA) at the Carl Hayden Bee Research Center. The EFA is divided into separate sections each of which are 2.5 m wide, 3.6 m long, and with a concave roof that is 4.3 m high at the top. Bees cannot fly between the sections and are not able to forage outside. One colony with a caged mated queen was placed in each section of the EFA. The queen was released within 24 h and began laying within 72 h of colony establishment.
Feeding colonies
Pollen feeding began when larvae were seen in EFA colonies. Ground pollen was placed on a tray outside of each colony, and the bees collected it in their corbiculae and brought it into the hive. Fresh pollen was added to the trays daily. During the first week of each experiment, all ten colonies in the EFA foraged on control pollen. Afterwards, five colonies were fed treatment pollen (either low or high dose) and five remained on control pollen. The bees consumed the pollen and used it to rear brood as it was collected, since there was little or no stored pollen in the hives during the study. After 21 days of feeding on either treatment or control pollen, nurse bees (five per colony) were sampled from all colonies. The nurse bees were identified as worker bees with their heads in cells containing larvae. The delay in nurse bee sampling assured us that the bees we collected were reared on and currently feeding exclusively on the pollen we supplied.
DNA extraction and 16S rRNA gene sequencing
For each bee, DNA was extracted from the entire digestive tract excluding the crop. Each gut was dissected directly into lysis buffer (180 μL TE/Triton X lysis buffer (20 mM Tris-HCl, 2 mM EDTA, 1.2 % Triton X-100, pH 8.0; 20 mg/mL lysozyme added immediately before use), and total genomic DNA was extracted using the GeneJet Genomic DNA Purification Kit (Fermentas) following the protocol for gram-positive bacteria. The V6–V8 variable region of the 16S ribosomal RNA (rRNA) gene sequence was PCR amplified from the total genomic DNA with the universal PCR primers 926F (5′-AAACTYAAAKGAATTGACGG-3′) and 1392R (5′-ACGGGCGGTGTGTRC-3′) using the HotStarTaq Plus Master Mix Kit (Qiagen, USA) and the following PCR conditions: 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s, 53 °C for 40 s, 72 °C for 1 min, and a final extension at 72 °C for 5 min. The products were visualized on a 2 % agarose gel and the 60 libraries (5 colonies × 2 treatments × 3 nurse bees × 2 experiments) were sequenced using the Illumina MiSeq protocol according to the manufacturer’s protocol. All sequence data were deposited in GenBank under Sequence Read Archive number SPR081297.
Pyrotagged sequence analysis
Sequence data were processed using QIIME version 1.9.0 (Caporaso et al. 2010b) and Mothur 1.35.0 (Schloss et al. 2009). Barcodes were extracted (extract_barcodes.py) and the libraries were split (split_libraries_fastq.py). Operational taxonomic units (OTUs) were picked at 97 % similarity with the UCLUST algorithm ((Edgar 2010); pick_otus.py). Representative sequences were selected for each OTU (pick_rep_set.py) and were aligned using PyNAST ((Caporaso et al. 2010a); align_seqs.py)). The aligned sequences were filtered (filter.seqs with no trump option). OTUs and representative sequences from the OTUs were selected from the filtered and aligned sequences as above. Chimeras were removed using the blast fragment option in identify_chimeric_seqs.py (Altschul et al. 1990). Taxonomy was assigned to the representative sequences using the RDP Naïve Bayesian Classifier ((Wang et al. 2007); in classify.seqs) and a manually constructed training set that contained sequences from the green genes 16S rRNA database (version gg_13_5_99), the RDP version 9 training set, and all full length honey bee-associated gut microbiota listed in NCBI (accessed July 2013) trimmed to the V6–V8 region. A table containing the number of sequences belonging to each OTU in each sequence library was constructed with the new taxonomy assignments (make_otu_table.py). OTUs containing fewer than two sequences per library and that were present in fewer than two libraries were discarded, as were those that were classified at less than 80 % confidence at the phylum level or that did not match a sequence in the NCBI nucleotide database at ≥97 % sequence similarity.
Analysis of microbial communities
For each experiment, we tested whether Pristine® impacted the relative abundance of taxa whose genomes contain high numbers of genes for carbohydrate digestion (Gilliamella apicola, Frischella perrara, Lactobacillus sp. Firm 4 and Firm 5, and Bifidobacterium sp. (Engel et al. 2012)). A generalized linear model with a normal distribution and identity link was used to analyze the data, where relative abundance was the response variable and treatment (Pristine® or control) and colony nested within treatment were the predictor variables. Each type of bacteria was analyzed separately. The relative abundances of the core honey bee gut taxa Acetobacteraceae sp. Alpha 2.1, Snodgrassella alvi, G. apicola, F. perrara, Lactobacillus sp. Firm 4 and Firm 5, and Bifidobacterium sp. found in other surveys of nurse guts (Martinson et al. 2011; Martinson et al. 2012; Moran et al. 2012) were also compared to the relative abundances of these taxa in nurse guts from the present study. For each treatment and for each experiment, the relative abundance of each taxon was calculated by totaling the number of sequences obtained belonging to that taxon divided by the total sequences obtained over all libraries subjected to that treatment.
Rarefaction curves (OTUs vs. sampling depth) were generated for each library (multiple_rarefaction.py) to determine the sampling depth. The slope of increase was effectively zero for each library. To calculate α- and β-diversity, a subsampling depth that maximized the number of libraries in the analysis was determined for all libraries in the experiment. Libraries with too few sequences were eliminated from the analyses. Only hives that had at least two acceptable libraries from two out of the three individuals sampled from that colony were included in the α- and β-diversity analyses.
To test whether taxon abundance within libraries differed due to fungicide treatment, Good’s coverage, the Chao index of species richness, and inverse Simpson diversity index were calculated for each rarefied library (alpha_diversity.py). The distribution of the Chao 1 estimate of species richness and Simpson diversity index was checked for normality and a t test was performed on these values. Tests were performed separately for each experiment.
To determine if fungicide treatment affected the distribution of bacterial taxa among libraries, representative sequences from each 97 % OTU were aligned (align.seqs) and filtered (filter.seqs, vertical=T, trump=.) and a phylogeny was constructed (clearcut; (Sheneman et al. 2006)). This phylogeny and the per-library abundance of each 97 % OTU were used in a jackknife analysis of unweighted and weighted UniFrac distances among libraries (jackknifed_beta_diversity.py) (Hamady et al. 2010; Lozupone and Knight 2005; Lozupone et al. 2011). The libraries were hierarchically clustered using UPGMA, and the support for the branches was calculated as the degree of jackknife support after subsampling ten times. If fungicide treatment strongly influenced the presence of bacterial taxa in individual nurses, we expected to see two clear, well-supported clades containing either fungicide-treated or untreated libraries.
A permutational multivariate analysis of variance (PERMANOVA) in the GUniFrac package (Chen et al. 2012) was also used to determine whether fungicide treatment affected the distribution of bacterial taxa among libraries. Representative sequences from each 97 % OTU and the per-library abundances of each 97 % OTU were analyzed using the phangorn (Schliep 2011), seqinr (Charif et al. 2005), ape (Paradis et al. 2004), and pegas (Paradis 2010) packages in R. A pairwise distance matrix was calculated from the sequence alignment, and a midpoint-rooted neighbor joining phylogeny was constructed. Rarefied estimates of the number of sequences per library per OTU were generated using the GUniFrac package (Chen et al. 2012) in R. Unweighted, weighted, variance adjusted weighted (VAW), and generalized (α = 0.5) pairwise distances were calculated (Chen et al. 2012; Hamady et al. 2010; Lozupone and Knight 2005; Lozupone et al. 2011). The PERMANOVA tested whether the combined UniFrac distances between libraries were significantly impacted by the fungicide treatment (Pristine® or the control) or colony nested within the fungicide treatment.
3 Results
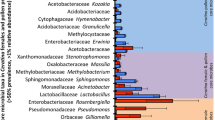
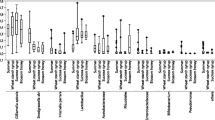
The individual libraries exhibited a considerable degree of individual variation in relative taxon abundance for both experiments (Online Resources 1, 2, and 3), with differences occurring among nurses sampled from the same treatment and within the same colony. Averaging among individuals within each colony gave a more consistent look at the among-colony variability and enabled comparisons between treatments and controls (Figure 1). The microbial communities from the guts of bees fed the fungicide-treated pollen and their associated control treatments were similar in overall composition, suggesting that Pristine® did not noticeably and consistently impact the distribution of nurse gut microbial communities.
The abundance of bacterial taxa in nurse honey bee guts. The pollen provided to colonies was treated with the fungicide Pristine® diluted in water at a concentration of either 1990 (experiment 1) or 11,410 (experiment 2) ppb. Water only was applied to the control pollen. Each bar represents the average relative abundance of bacteria in the guts of three nurse worker honey bees (number of sequences for that taxon/total sequences for the library). Individual abundances are presented in Online Resources 1, 2, and 3. Libraries marked with an asterisk (*) were not included in calculations of α- or β-diversity due to small sample size.
Despite the fact that Pristine® did not have a consistent impact on the nurse gut microbial communities for both trials, there were some interesting fungicide-related effects (Figure 1 and Online Resources 2 and 3) within each experiment. In the first experiment (low dose), the general linear model (GLM) analyses revealed that the abundance of Gilliamella sp. sequences in the sequencing libraries was reduced by Pristine® (mean relative abundance in control colonies = 8.9 ± 3.5 % s.e., mean relative abundance in Pristine®-treated colonies = 2.2 ± 0.5 % s.e.; Χ 2 1 = 4.71, P = 0.03). All other potentially carbohydrate-degrading taxa were unaffected by the treatment. Colony variability nested within treatment also did not impact the relative abundances of these carbohydrate-degrading taxa. In bees fed the high dose of Pristine®, the GLM analysis indicated a significant increase in the number of Lactobacillus sp. Firm 4 (mean relative abundance in control colonies = 0.6 ± 0.3 % s.e., mean relative abundance in Pristine®-treated colonies = 6.6 ± 3.2 % s.e.; Χ 2 1 = 5.89, P = 0.015) and Lactobacillus sp. Firm 5 (mean relative abundance in control colonies = 1.6 ± 0.8 % s.e., mean relative abundance in Pristine®-treated colonies = 13.7 ± 6.4 % s.e.; Χ 2 1 = 5.75, P = 0.016) sequences in the sequencing libraries. The among-colony variability also was a significant predictor of the abundances of Lactobacillus sp. Firm 4 (Χ 2 8 = 34.4, P < 0.0001), Lactobacillus sp. Firm 5 (Χ 2 8 = 33.4, P < 0.0001), and F. perrara (Χ 2 8 = 19.7, P = 0.011) sequences in the sequencing libraries.
The individual libraries were averaged within colonies to compare the results of the present study to previous studies of nurse gut bacterial communities (Martinson et al. 2011; Martinson et al. 2012; Moran et al. 2012). The core honey bee gut bacteria (Alpha 2.1, S. alvi, G. apicola, F. perrara, Lactobacillus sp. Firm 4 and Firm 5, and Bifidobacterium sp.) were present in almost all samples in both experiments (Figure 1, Table I, and Online Resources 1, 2, and 3). In bees fed the low dose of Pristine®, all core gut taxa were within the ranges of proportions typically found in hive bees with the exception of S. alvi and G. apicola, which were lower than reported in other studies (Table I). The levels of non-core gut microbiota were higher than reported previously. A large proportion of non-core sequences (10 % of total sequences in experiment) shared ≥97 % sequence identity with Bartonella apis (Kesnerova et al. 2015). B. apis was especially prevalent in two individuals collected from one control (77 % of total sequences for that individual) and one Pristine®-treated colony (74 % of total sequences for that individual; Online Resources 1 and 2).
In bees fed the high dose of Pristine®, levels of Acetobacteraceae sp. Alpha 2.1, F. perrara, Lactobacillus sp. Firm 4, and Bifidobacterium were within or just slightly below (0.6 %) the expected ranges based on previous studies. However, levels of S. alvi, G. apicola, and Lactobacillus sp. Firm 5 were greatly reduced in both the treatment and control libraries (Table I) compared to previous studies. Many non-core taxa also were abundant, especially Parasaccharibacter apium (25 % of total sequences), non-Firm 4 and Firm 5 Lactobacillus sp. (34 % of total sequences), and Enterococcaceae (24 % of total sequences). Several individuals exhibited higher than expected levels of Paenibacillus larvae, especially one individual from colony 4 fed the control-treated pollen (Online Resource 3). In the remainder of colonies, levels of P. larvae were less than 1 % of the total sequences sampled in all of the colonies. Representative sequences for the 97 % OTUs from both experiments are provided in the Online Resources 4 and 5.
A subsampling depth of 13,821 (low dose) and 2815 (high dose) maximized the number of libraries that could be reliably analyzed. We excluded two colonies fed the low dose of Pristine® (hives 4 and 10) and one fed the high dose (hive 5) from α- and β-diversity analyses due to low sampling depth (Figure 1). In addition, one individual from colony 7 and one from colony 9 were also excluded from the low-dose experiment (Online Resource 2) and one individual from colony 2 and one from colony 7 were excluded from the high-dose experiment (Online Resource 3). The average coverage of the remaining rarefied libraries was above 99 % for both experiments.
Pristine® significantly reduced the number of OTUs in each library in the low-dose treatment (1990 ppb; average number of OTUs in control 325.75 ± 37.04 s.e., average number of OTUs in treated 151.25 ± 10.76 s.e.; t 6 = −4.52; P = 0.004), but not in the high dose (Table II). The diversity of bacteria within libraries, as measured by the Simpson diversity index, was not impacted by Pristine® at either high or low doses (Table II). The presence of fungicide did not significantly impact the distribution of bacterial taxa among the libraries. In both experiments, phylogenies based on both weighted and unweighted UniFrac distances exhibited well-supported nodes, but no clear separation between clades due to the fungicide treatment (Online Resource 4). Libraries from the same colony rarely grouped within the same clade; instead, they often were more related to individual libraries from different colonies and treatments. The PERMANOVA analysis revealed no significant effect of the fungicide or colony nested within the fungicide treatment on the distribution of bacteria among libraries.
4 Discussion
The core taxa that normally comprise honey bee gut bacterial communities were present to varying degrees in most nurse bees. Non-core bacteria also were present, sometimes at higher levels than expected based on previous studies. The abundance of the putative carbohydrate-degrading genera Gilliamella, Frischella, Bifidobacterium, and the Lactobacillus sp. Firms 4 and 5 was impacted by Pristine® in different ways depending on the experiment. Bacterial species richness was significantly reduced at the low dose of Pristine®, but not at the high dose. Diversity was not impacted by the treatment in either experiment. There were notable differences between experiments that might have been due to differences in pollen consumption between high and low doses of Pristine® (experiment 2) (DeGrandi-Hoffman et al. 2015) or environmental and dietary factors that can affect the gut microbiome in honey bees and other organisms (Ludvigsen et al. 2015).
Though strobilurin fungicides have bactericidal properties (Bartlett et al. 2002), gut microbial communities in nurse bees were not consistently altered by feeding on pollen with added Pristine®. The number of bacterial OTUs decreased due to the application of 1990 ppb but not 11,410 ppb of Pristine®. Gilliamella sp. abundance was negatively impacted by the fungicide in the low-dose experiment but not in the high dose. Conversely, the abundance of Lactobacillus sp. Firm 4 and Firm 5 was positively impacted by Pristine® in the high but not the low-dose experiments. There are at least three possible explanations for the disparate results we obtained from this study. The simplest is that the exposure to Pristine® was too low to affect gut microbial communities. We applied field-relevant rates of Pristine® to the pollen, but the actual exposure to the bees might have been considerably lower. A previous study reported significant reductions in the concentration of components of Pristine® in bee bread and nurse bees relative to amounts in the pollen they were fed (DeGrandi-Hoffman et al. 2013). The reductions might be due to dilution with nectar (in this study sugar syrup) while packing the pollen in corbiculae or comb cells, or possibly from biotransformation of the compounds following consumption (Mullin et al. 2010). There also is evidence that the palatability of pollen is reduced when Pristine® is added, so bees may have actually consumed less of the high-dose pollen (DeGrandi-Hoffman et al. 2015; Campbell et al. 2016). A second explanation is that the greater than expected variability in the composition of gut microbial communities in nurse bees from the same colony and between years may have obscured the effects of Pristine®. Different outcomes may have occurred if both the low- and high-dose experiments were performed in the same year using bees from the same source colonies. We might have been better able to capture the variability among nurses within colonies if more were sampled per colony. A final explanation is that recurrent consumption of pollens containing fungicides may have led to resistance in the gut microbiota. Fungicides are commonly detected in the pollen from managed colonies (Mullin et al. 2010). Whether fungicides impose a strong enough fitness cost to the microbes and whether this leads to selection of resistant strains is not clear. However, species of Bacillus isolated from soils exposed to high doses of azoxystrobin show resistance to this fungicide (Bacmaga et al. 2015). There also is evidence that gut microbiota in honey bees can acquire resistance to antibiotics, reflecting the frequent use for foulbrood control (Tian et al. 2012).
The expected species composition of the gut microbiota was present in most nurse bees from both experiments, but their abundances were somewhat different from those reported in other studies. In the first experiment, G. apicola was less abundant than expected and B. apis was highly abundant in two individuals, one from a control and another from a treatment colony. Bartonella is sporadic in its occurrence in pyrosequencing libraries derived from pooled (Corby-Harris et al. 2014a; Martinson et al. 2011; Martinson et al. 2012; Moran et al. 2012; Sabree et al. 2012) and individual bee samples (Ahn et al. 2012; Corby-Harris et al. 2014a, and the present study), suggesting that B. apis is transmitted with only low or medium fidelity. In the second experiment, we observed a large number of non-core bacteria, particularly P. apium, non-Firm 4 and Firm 5 Lactobacillus sp., Enterococcaceae, and the pathogen P. larvae. P. apium is found at lower levels in the gut (Corby-Harris et al. 2014a; Martinson et al. 2011; Martinson et al. 2012; Moran et al. 2012; Sabree et al. 2012) and is typically more abundant in hive food stores (Anderson et al. 2014), queens (Kapheim et al. 2015; Tarpy et al. 2015), larvae (Vojvodic et al. 2013), and in the hypopharyngeal glands and crops of nurses (Corby-Harris et al. 2014b). Because P. apium is relatively abundant in food stores, hypopharyngeal glands, and crop, one possibility is that the guts displaying high P. apium titers were recently inoculated through either pollen feeding or trophallaxis.
There were considerable differences in the composition of the microbial communities between the two experiments. These differences may be due to the composition and nutritional value of the pollen used for feeding the bees. We fed the bees polyfloral mixes of pollen that were collected during the year when the experiment was conducted, so the species composition probably differed. There is evidence that bee bread communities (Anderson et al. 2014) and midgut/pyloric (Ludvigsen et al. 2015) bacterial communities fluctuate with pollen source and season. Gut bacterial communities also can differ among colonies (Hroncova et al. 2015). Bees for each of our experiments were sourced from different sets of colonies, and this might also have contributed to the differences we detected in gut microbiota between the experiments. It is important to note that although the use of the enclosed flight arena (EFA) permitted us to rear colonies on only the pollen we provided, this is not the normal foraging environment for honey bees. Bumblebees reared inside enclosures have a drastically different gut microbiota compared to bees found in nature (Meeus et al. 2015; Parmentier et al. 2015). Though we detected core taxa normally comprising honey bee gut bacterial communities, it is possible that the sporadic abundance of normally rare taxa and underrepresentation of taxa that are normally more abundant was due to housing colonies in an EFA. For example, the honey bee pathogen P. larvae was detectable in all of the colonies from the second experiment and reached very high levels in one of the hives. If the colonies used for this experiment had a low level of infection prior to being confined in the EFA, this confinement combined with limited access to the outside environment may have contributed to the high incidences of P. larvae in these hives.
Previous studies focused on the effects of Pristine® on nutrient acquisition, and metabolism found reduced consumption and digestion of pollen contaminated with this fungicide (Campbell et al. 2016). Results from the current study indicate that reduced digestion may not be related to changes in the gut microbiota, but rather to the direct effects on the bees, such as respiratory inhibition and reductions in ATP synthesis. Further studies are needed to confirm these effects.
References
Ahn J-H, Hong I-P, Bok J-I, Kim B-Y, Song J, et al. (2012) Pyrosequencing analysis of the bacterial communities in the guts of honey bees Apis cerana and Apis mellifera in Korea. J Microbiol 50:735–745
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J. Mol. biol. 215:403–410
Anderson K.E., Sheehan, T., Mott, B., Maes, P., Snyder, L., Schwan, M.R., Walton, A., Jones, B., Corby-Harris, V. (2013) Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PloS one 8:e83125 doi:10.1371/journal.pone.0083125
Anderson KE, Carroll MJ, Sheehan T, Mott BM, Maes P, Corby-Harris V (2014) Hive-stored pollen of honey bees: many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol. ecol. 23:5904–5917
Anderson, K.E., Rodrigues, P.A.P., Mott, B.M., Maes, P., Corby-Harris, V. (2015) Ecological succession in the honey bee gut: shift in Lactobacillus strain dominance during early adult development. Microb. Ecol., 1–12
Anke T (1995) The antifungal strobilurins and their possible ecological role Canadian. J. Bot. 73:940–945
Arnoult D, Carneiro L, Tattoli I, Girardin SE (2009) The role of mitochondria in cellular defense against microbial infection. Seminars in immunology 21:223–232
Babendreier D, Joller D, Romeis J, Bigler F, Widmer F (2007) Bacterial community structures in honeybee intestines and their response to two insecticidal proteins. FEMS microbiol. ecol. 59:600–610
Bacmaga M, Kucharski J, Wyszkowska J (2015) Microbial and enzymatic activity of soil contaminated with azoxystrobin. Environ. monit. assess. 187:615
Bartlett DW, Clough JM, Godwin JR, Hall AA, Hamer M, Parr-Dobrzanski B (2002) The strobilurin fungicides. Pest Manag. Sci 58:649–662
Blacquiere T, Smagghe G, van Gestel CA, Mommaerts V (2012) Neonicotinoids in bees: a review on concentrations, side-effects and risk assessment. Ecotoxicology 21:973–992
Campbell JB, Nath R, Gadau J, Fox T, DeGrandi-Hoffman G, Harrison JF (2016) The fungicide Pristine® inhibits mitochondrial function in vitro but not flight metabolic rates in honey bees. J. Insect Physiol. 86:11–16
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010a) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
Caporaso JG, Kuczynski J., Stombaugh, J., Bittinger, K., Bushman, F. et al. (2010b) QIIME allows analysis of high-throughput community sequencing data. Nat. Meth. 7:335–336
Charif D, Thioulouse J, Lobry JR, Perriere G (2005) Online synonymous codon usage analyses with the ade4 and seqinR packages. Bioinformatics 21:545–547
Chen J., Bittinger, K., Charlson, E., Hoffmann, C., Lewis, J., Wu, G.D., Collman, R.G., Bushman, F.D., Li, H. et al. (2012) Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28:2106–2113
Corby-Harris V, Maes P, Anderson KE (2014a) The bacterial communities associated with honey bee Apis mellifera foragers. PloS one 9:e95056 doi:10.1371/journal.pone.0095056
Corby-Harris V, Snyder LA, Schwan MR, Maes P, McFrederick QS, Anderson KE (2014b) Origin and effect of Alpha 2.2 Acetobacteraceae in honey bee larvae and description of Parasaccharibacter apium gen. nov., sp. nov. Appl. environ. microbiol. 80:7460–7472
DeGrandi-Hoffman G, Chen Y, Simonds R (2013) The Effects of Pesticides on Queen Rearing and Virus Titers in Honey Bees (Apis mellifera L.). Insects 4:71–89
DeGrandi-Hoffman G, Chen Y, Watkins Dejong E, Chambers ML, Hidalgo G (2015) Effects of Oral Exposure to Fungicides on Honey Bee Nutrition and Virus Levels. J. econ. entomol. 108:2518–2528
Dively GP, Embrey MS, Kamel A, Hawthorne DJ, Pettis JS (2015) Assessment of chronic sublethal effects of imidacloprid on honey bee colony health. PloS one 10:e0118748 doi:10.1371/journal.pone.0118748
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Engel P, Martinson VG, Moran NA (2012) Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A 109:11002–11007
Gill RJ, Ramos-Rodriguez O, Raine NE (2012) Combined pesticide exposure severely affects individual- and colony-level traits in bees. Nature 491:105–108
Gottesman S, Maurizi MR (1992) Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. rev. 56:592–621
Hamady M, Lozupone C, Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. The ISME J. 4:17–27
Hroncova Z., Havlik, J., Killer, J., Doskocil, I., Tyl, J., Kamler, M., Titera, D., Hakl, J., Mrazek, J., Bunesova, V., et al. (2015) Variation in honey bee gut microbial diversity affected by ontogenetic stage, age and geographic location PloS one 10:e0118707 doi:10.1371/journal.pone.0118707
Janousek CN, Gubler WD (2010) Control of brown rot and shot hole in almond: 2009 field trial. (https://escholarship.org/uc/item/7zh4h834)
Kapheim KM, Rao VD, Yeoman CJ, Wilson BA, White BA, Goldenfeld N, Robinson GE (2015) Caste-specific differences in hindgut microbial communities of honey bees (Apis mellifera). PloS one 10:e0123911 doi:10.1371/journal.pone.0123911
Kesnerova L, Moritz R, Engel P (2015) Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. doi:10.1099/ijsem.0.000736
Koch H, Schmid-Hempel P (2011) Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci U S A 108:19288–19292
Kuhn PJ (1984) Mode of action of carboximides. Symp Ser Br Mycol Soc 9:155–183
Lee FJ, Rusch DB, Stewart FJ, Mattila HR, Newton IL (2015a) Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ. microbiol. 17:796–815
Lee K.V., Steinhauer N., Rennich K., Wilson M.E., Tarpy D. R., Caron D. M., Rose R., Delaplane K.S., Baylis K., Lengerich E. J., et al. (2015b) A national survey of managed honey bee 2013–2014 annual colony losses in the USA. Apidologie 46:292–305
Legard DE, Xiao CL, Mertely JC, Chandler CK (2001) Management of Botrytis Fruit Rot in Annual Winter Strawberry Using Captan, Thiram, and Iprodione. Plant Dis. 85:31–39
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235
Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R (2011) UniFrac: an effective distance metric for microbial community comparison. The ISME journal 5:169–172
Ludvigsen J, Rangberg A, Avershina E, Sekelja M, Kreibich C, Amdam G, Rudi K (2015) Shifts in the Midgut/Pyloric Microbiota Composition within a Honey Bee Apiary throughout a Season. Microbes and environments/JSME 30:235–244
Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA (2011) A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. ecol. 20:619–628
Martinson VG, Moy J, Moran NA (2012) Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. environ. microbiol. 78:2830–2840
Meeus I., Parmentier, L., Billiet, A., Maebe, K., Van Nieuwerburgh, F., Deforce, D., Wackers, F., Vandamme, P., Smagghe, G. (2015) 16S rRNA Amplicon Sequencing Demonstrates that Indoor-Reared Bumblebees (Bombus terrestris) Harbor a Core Subset of Bacteria Normally Associated with the Wild Host. PloS one 10:e0125152 doi:10.1371/journal.pone.0125152
Moran NA, Hansen AK, Powell JE, Sabree ZL (2012) Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PloS one 7:e36393 doi:10.1371/journal.pone.0036393
Mullin CA, Frazier M, Frazier JL, Ashcraft S, Simonds R, vanEngelsdorp D, Pettis JS (2010) High Levels of Miticides and Agrochemicals in North American Apiaries: Implications for Honey Bee Health. PloS one 5:e9754 doi:10.1371/journal.pone.0009754
Paradis E (2010) pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420
Paradis E, Claude J, Strimmer K (2004) APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20:289–290
Parmentier, L., Meeus, I., Mosallanejad, H., Graaf, D.C., Smagghe, G. (2015) Plasticity in the gut microbial community and uptake of Enterobacteriaceae (Gammaproteobacteria) in Bombus terrestris bumblebees’ nests when reared indoors and moved to an outdoor environment. Apidologie, 1–14
Pettis JS, Lichtenberg EM, Andree M, Stitzinger J, Rose R, Vanengelsdorp D (2013) Crop pollination exposes honey bees to pesticides which alters their susceptibility to the gut pathogen Nosema ceranae. PloS one 8:e70182 doi:10.1371/journal.pone.0070182
Sabree ZL, Hansen AK, Moran NA (2012) Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PloS one 7:e41250 doi:10.1371/journal.pone.0041250
Saraiva M.A., Pegoraro Zemolin, A.P., Franco, J.L., Boldo, J.T., Stefenon, V.M., Triplett, E.W., de Oliveira Camargo, F.A., Wurdig Roesch, L.F. (2015) Relationship between honeybee nutrition and their microbial communities. Antonie van Leeuwenhoek Int. J. Gen. Mol. Biol. 107 (4):921–933
Schliep KP (2011) phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593
Schloss PD et al. (2009) Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. environ. microbiology 75:7537–7541
Sheneman L, Evans J, Foster JA (2006) Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics 22:2823–2824
Simon-Delso N, SanMartin G, Bruneau E, Minsart L-A, Mouret C, Hautier L (2014) Honeybee colony disorder in crop areas: the role of pesticides and viruses. PLoS ONE 9:e103073. doi:10.1371/journal.pone.0103073
Tarpy DR, Mattila HR, Newton ILG (2015) Characterization of the honey bee microbiome throughout the queen-rearing process. Appl. environ. microbiol. doi:10.1128/aem.00307-15
Tian, B., Fadhil, N.H., Powell, J.E., Kwong, W.K., Moran, N.A. (2012) Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 3. doi:10.1128/mBio.00377–12
Vojvodic S, Rehan SM, Anderson KE (2013) Microbial Gut Diversity of Africanized and European Honey Bee Larval Instars. PloS one 8:e72106 doi:10.1371/journal.pone.0072106
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. environ. microbiol. 73:5261–5267
Yang C, Hamel C, Vujanovic V, Gan Y (2011) Fungicide: Modes of Action and Possible Impact on Nontarget Microorganisms. ISRN Ecology 2011:8 doi:10.5402/2011/130289
Yoshimura MA, Luo Y, Ma Z, Michailides TJ (2004) Sensitivity of Monilinia fructicola from Stone Fruit to Thiophanate-Methyl, Iprodione, and Tebuconazole. Plant Disease 88:373–378
Acknowledgments
The authors thank Roger Simonds for pesticide analysis, and Kirsten Shoshanna Traynor, Kirk Anderson, and two anonymous reviewers for their comments on a previous version of the manuscript. This research was funded by the United States Department of Agriculture (USDA) Agricultural Research Service (ARS). The ARS/USDA is an equal opportunity employer and provider.
Authors’ contributions
GD-H experimental design, data analysis, and co-wrote the manuscript with C-H microbiome and bioinformatics analyses; other co-authors establish and maintain colonies, pollen diets, and data collection
Author information
Authors and Affiliations
Corresponding author
Additional information
Manuscript editor: Klaus Hartfelder
Les communautés microbiennes du tube digestif de l’abeille résistent au fongicide Pristine® consommé via le pollen
respiration / nutrition / Apis mellifera / boscalide / pyraclostrobine / microbiome
Die Microbiotagemeinschaften im Darm der Honigbiene sind robust gegen das mit Pollen aufgenommene Fungizid Pristine®
atmung / ernährung / boscalid / pyraclostrobin / microbiom
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
DeGrandi-Hoffman, G., Corby-Harris, V., DeJong, E.W. et al. Honey bee gut microbial communities are robust to the fungicide Pristine® consumed in pollen. Apidologie 48, 340–352 (2017). https://doi.org/10.1007/s13592-016-0478-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-016-0478-y