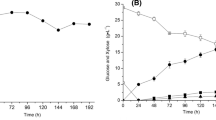
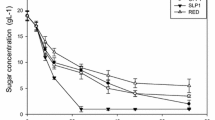
Abstract
The aims of this work were to evaluate the ability of furaldehyde-tolerant yeast strains Saccharomyces cerevisiae P6H9 and Wickerhamomyces anomalus WA-HF5.5 and their cofermentations and to convert soybean and rice hull hydrolysates into ethanol and xylitol. In batch shaker cultures, the strains showed the ability to tolerate high osmotic pressure (1918 mOsmkg−1), completely depleting furaldehyde in the first 12 h of cultivations, while converting the hydrolysate sugars into ethanol. Highest ethanol yields of 0.37 g g−1 and productivity of 0.31 g L−1 h−1 were obtained in the cofermentation using rice hull hydrolysate as substrate. The concentration of sugars in soybean hull hydrolysate proved to be inadequate as substrate for the cultivation of these strains, showing a low ethanol productivity of 0.08 g L−1 h−1. Bioreactor cultivations of S. cerevisiae on rice hull hydrolysate under anaerobiosis showed a relatively high ethanol productivity of 6.7 g L−1 h−1, whereas the bioreactor cofermentation produced xylitol to yields of 0.86 g g−1 under conditions of oxygen limitation.


Similar content being viewed by others
References
Chen Y (2011) Development and application of co-culture for ethanol production by co-fermentation of glucose and xylose: a systematic review. J Ind Microbiol Biotechnol 38:581–597
Keating J, Robinson J, Cotta M, Saddler J, Mansfield S (2004) An ethanologenic yeast exhibiting unusual metabolism in the fermentation of lignocellulosic hexose sugars. J Ind Microbiol Biotechnol 31:235–244
Almeida JR, Röder A, Modig T, Laadan B, Lidén G, Gorwa-Grauslund MF (2008) NADH- vs NADPH-coupled reduction of 5-hydroxymethyl furfural (HMF) and its implications on product distribution in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 78:939–945
da Cunha-Pereira F, Hickert LR, Sehnem NT, de Souza-Cruz PB, Rosa CA, Ayub MA (2011) Conversion of sugars present in rice hull hydrolysates into ethanol by Spathaspora arborariae, Saccharomyces cerevisiae, and their co-fermentations. Bioresour Technol 102:4218–4225
Margeot A, Hahn-Hagerdal B, Edlund M, Slade R, Monot F (2009) New improvements for lignocellulosic ethanol. Curr Opin Biotechnol 20:372–380
Yadav K, Naseeruddin S, Prashanthi G, Sateesh L, Rao L (2011) Bioethanol fermentation of concentrated rice straw hydrolysate using co-culture of Saccharomyces cerevisiae and Pichia stipitis. Bioresource Technol 102:6473–6478
Ko C, Chiu P, Yang C, Chang K (2008) Xylitol conversion by fermentation using five yeast strains and polyelectrolyte-assisted ultrafiltration. Biotechnol Lett 30:81–86
Schirmer-Michel A, Flores S, Hertz P, Matos G, Ayub M (2008) Production of ethanol from soybean hull hydrolysate by osmotolerant Candida guilliermondii NRRL Y-2075. Bioresource Technol 99:2898–2904
Tao N, Gao Y, Liu Y (2011) Isolation and characterization of a Pichia anomala strain: a promising candidate for bioethanol production. Braz J Microbiol 42:668–675
Zhao L, Zhang X, Tan T (2008) Influence of various glucose/xylose mixtures on ethanol production by Pachysolen tannophilus. Biomass Bioenergy 32:1156–1161
Vandeska E, Amartey S, Kuzmanova S, Jeffries T (1995) Effects of environmental-conditions on production of xylitol by Candida-boidinii. World J Microbiol Biotechnol 11:213–218
Zhao X, Bai F (2009) Mechanisms of yeast stress tolerance and its manipulation for efficient fuel ethanol production. J Biotechnol 144:23–30
Allen S, Clark W, McCaffery J, Cai Z, Lanctot A, Slininger P, Liu Z, Gorsich S (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:2–10
Sehnem NT, da Silva MA, Leite FC, de Barros PW, de Morais MA, Ayub MA (2013) 5-hydroxymethylfurfural induces ADH7 and ARI1 expression in tolerant industrial Saccharomyces cerevisiae strain P6H9 during bioethanol production. Bioresour Technol 133:190–196
Brat D, Boles E, Wiedemann B (2009) Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl Environ Microbiol 75:2304–2311
Kuyper M, Harhangi H, Stave A, Winkler A, Jetten M, de Laat W, den Ridder J, Op den Camp H, van Dijken J, Pronk J (2003) High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res 4:69–78
Petersson A, Almeida J, Modig T, Karhumaa K, Hahn-Hagerdal B, Gorwa-Grauslund M, Liden G (2006) A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23:455–464
Delgenes J, Laplace J, Moletta R, Navarro J (1996) Comparative study of separated fermentations and cofermentation processes to produce ethanol from hardwood derived hydrolysates. Biomass Bioenergy 11:353–360
Qian M, Tian S, Li X, Zhang J, Pan Y, Yang X (2006) Ethanol production from dilute-acid softwood hydrolysate by co-culture. Appl Biochem Biotechnol 134:273–283
Rouhollah H, Iraj N, Giti E, Sorah A (2007) Mixed sugar fermentation by Pichia stipitis, Sacharomyces cerevisiaea, and an isolated xylose-fermenting Kluyveromyces marxianus and their cocultures. Afr J Biotechnol 6:1110–1114
Liu ZL, Ma M, Song M (2009) Evolutionarily engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol Gen Genomics 282:233–244
Passoth V, Olstorpe M, Schnürer J (2011) Past, present and future research directions with Pichia anomala. Antonie Van Leeuwenhoek 99:121–125
Cassales A, de Souza-Cruz P, Rech R, Ayub M (2011) Optimization of soybean hull acid hydrolysis and its characterization as a potential substrate for bioprocessing. Biomass Bioenergy 35:4675–4683
Dien BS, Kurtzman CP, Saha BC, Bothast RJ (1996) Screening for L-arabinose fermenting yeasts. Appl Biochem Biotechnol 57:233–242
Becker J, Boles E (2003) A modified Saccharomyces cerevisiae strain that consumes L-arabinose and produces ethanol. Appl Environ Microbiol 69:4144–4150
Hickert LR, da Cunha-Pereira F, de Souza-Cruz PB, Rosa CA, Ayub MA (2013) Ethanogenic fermentation of co-cultures of Candida shehatae HM 52.2 and Saccharomyces cerevisiae ICV D254 in synthetic medium and rice hull hydrolysate. Bioresour Technol 131:508–514
Dhar R, Sägesser R, Weikert C, Yuan J, Wagner A (2011) Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J Evol Biol 24:1135–1153
Pandey G, Yoshikawa K, Hirasawa T, Nagahisa K, Katakura Y, Furusawa C, Shimizu H, Shioya S (2007) Extracting the hidden features in saline osmotic tolerance in Saccharomyces cerevisiae from DNA microarray data using the self-organizing map: biosynthesis of amino acids. Appl Microbiol Biotechnol 75:415–426
Garay-Arroyo A, Covarrubias AA, Clark I, Niño I, Gosset G, Martinez A (2004) Response to different environmental stress conditions of industrial and laboratory Saccharomyces cerevisiae strains. Appl Microbiol Biotechnol 63:734–741
Almeida J, Modig T, Petersson A, Hahn-Hagerdal B, Liden G, Gorwa-Grauslund M (2007) Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol 82:340–349
Martin C, Marcet M, Almazan O, Jonsson L (2007) Adaptation of a recombinant xylose-utilizing Saccharomyces cerevisiae strain to a sugarcane bagasse hydrolysate with high content of fermentation inhibitors. Bioresource Technol 98:1767–1773
Mussatto SI, Dragone G, Roberto IC (2005) Influence of the toxic compounds present in brewer's spent grain hemicellulosic hydrolysate on xylose-to-xylitol bioconversion by Candida guilliermondii. Process Biochem 40:3801–3806
Wahlbom CF, Hahn-Hägerdal B (2002) Furfural, 5-hydroxymethyl furfural, and acetoin act as external electron acceptors during anaerobic fermentation of xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng 78:172–178
Guo C, He P, Lu D, Shen A, Jiang N (2006) Cloning and molecular characterization of a gene coding D-xylulokinase (CmXYL3) from Candida maltosa. J Appl Microbiol 101:139–150
de Smidt O, du Preez J, Albertyn J (2008) The alcohol dehydrogenases of Saccharomyces cerevisiae: a comprehensive review. FEMS Yeast Res 8:967–978
Acknowledgements
The authors wish to thank the Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq), Coordenação do Aperfeiçoamento de Pessoal do Ensino Superior.
(CAPES) and Fundação do Amparo a Pesquisa do Estado do Rio Grande do Sul (FAPERGS) for their financial support for the project and scholarships.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sehnem, N.T., Hickert, L.R., da Cunha-Pereira, F. et al. Bioconversion of soybean and rice hull hydrolysates into ethanol and xylitol by furaldehyde-tolerant strains of Saccharomyces cerevisiae, Wickerhamomyces anomalus, and their cofermentations. Biomass Conv. Bioref. 7, 199–206 (2017). https://doi.org/10.1007/s13399-016-0224-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-016-0224-8