Abstract
The smaller tea tortrix, Adoxophyes honmai Yasuda (Lepidoptera: Tortricidae), is one of the main insect pests of tea, Camellia sinensis Kuntz, in Japan. Recently, A. honmai has developed a high resistance to diacylhydrazine analog insect growth regulators, such as tebufenozide, in Shizuoka Prefecture. Previously, we identified a point mutation (A415V) in the ecdysone receptor gene (EcR), a candidate factor responsible for tebufenozide resistance. In this study, we have developed a molecular method of diagnosis to detect the EcR A415V mutation by polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP). This method was confirmed to be successfully applicable to larvae reared in the laboratory and adults collected by pheromone traps in the field. The appearance ratio of the resistant allele in the A. honmai populations from various Japanese districts examined by the method revealed a high correlation with the magnitude of tebufenozide resistance. These results verified that the A415V mutation is the principal factor responsible for tebufenozide resistance and the PCR–RFLP method may be used as a reliable and convenient tool for monitoring tebufenozide resistance in the field.
Similar content being viewed by others
Introduction
The smaller tea tortrix, Adoxophyes honmai Yasuda (Lepidoptera: Tortricidae), is a polyphagous insect pest infesting many agricultural crops, including tea, citrus, rose, and grape [9]. Adoxophyes honmai has 4–5 generations per year in the tea fields of Shizuoka Prefecture, which produces the largest quantity of tea in Japan [20]. Larvae prefer feeding on leaf buds and young leaves to old leaves [12]. Therefore, outbreaks of this pest cause severe damage to tea leaves, developmental delay of tea shoots, reductions in tea yield, and decreased quality in tea manufacturing.
Various insecticides have been used for the control of A. honmai in Shizuoka Prefecture to date. However, A. honmai has already developed resistance to many insecticides, including carbamates [13], organophosphates, synthetic pyrethroids, and benzoylurea analog insect growth regulators (IGRs) [7]. In addition, the development of high levels of resistance to diacylhydrazine (DAH) analog IGRs and diamide insecticides has been recently reported [19, 20].
DAH analog IGRs, including tebufenozide, chromafenozide, and methoxyfenozide are nonsteroidal ecdysteroid agonists. They exhibit insecticidal activity by binding to the target protein, ecdysone receptor, and promoting aberrant ecdysis in the larvae of lepidopteran insect pests [11, 24, 25]. Tebufenozide has been used as a popular insecticide for the control of major lepidopteran pests, A. honmai and the oriental tea tortrix Homona magnanima Diakonoff (Lepidoptera: Tortricidae), in the tea fields of Shizuoka Prefecture since 1993 [7]. However, A. honmai quickly developed resistance to tebufenozide, where the resistant ratio of a population collected in the Makinohara region of Shizuoka Prefecture in 2004 was 57.7-fold compared to a susceptible strain, and reached 1,528-fold in 2008 [19].
In addition to A. honmai, the development of tebufenozide resistance has been reported in field populations of the apple pandemis leafroller moth Pandemis pyrusana Kearfott (Lepidoptera: Tortricidae) [3], the codling moth Cydia pomonella L. (Lepidoptera: Tortricidae) [5], and the oblique-banded leafroller moth Choristoneura rosaceana Harris (Lepidoptera: Tortricidae) [3, 15], and in laboratory-selected strains of the diamondback moth Plutella xylostella L. (Lepidoptera: Plutellidae) [2], the beet armyworm Spodoptera exigua Hübner (Lepidoptera: Noctuidae) [6], and the greenheaded leafroller Planotortrix octo Dugdale (Lepidoptera: Tortridae) [10, 23]. However, the mechanisms of tebufenozide resistance remain largely unknown in these species.
Genetic studies have revealed that tebufenozide resistance in A. honmai is incompletely dominant and is associated with multiple autosomal genes [21]. More recently, by next-generation sequencing analyses, we have identified two types of candidate genes that are putatively responsible for the tebufenozide resistance: (1) ecdysone receptor gene (EcR) that encodes the target molecule of tebufenozide, and (2) cytochrome P450 monooxygenase genes (CYP9As) that encode metabolic enzymes [18]. In resistant strains, a single nucleotide polymorphism (SNP) causing an amino acid substitution of alanine (A) at position 415 to valine (V) was found in the ligand binding domain of EcR. These amino acids adjoin an amino acid residue critical for DAH binding. Moreover, biochemical analyses using a recombinant EcR protein showed that the A415V mutation resulted in a 5-fold decrease in the binding affinity for tebufenozide. Meanwhile, the expression level of CYP9A genes were high in resistant strains and moderately correlated with the magnitude of tebufenozide-resistance. Thus, both the desensitization of target protein (EcR) and the enhancement of detoxification activity by overexpressed CYP9As are relevant to the resistant mechanism [18]. However, it remains unclear to what extent the EcR mutation contributes to tebufenozide resistance in A. honmai in the field.
Here, we have developed a molecular diagnostic method targeting the A415V mutation in EcR. Using this method, we elucidated the relationship between the A415V mutation and the magnitude of tebufenozide resistance in various A. honmai field populations. Accordingly, we revealed that the A415V mutation is indeed a principal factor in tebufenozide resistance and this method is useful for field-monitoring of tebufenozide-resistance in A. honmai.
Materials and methods
Insects
Larval or adult A. honmai were collected in the tea fields of Shizuoka, Mie, Kyoto, Saitama, Ibaraki, Kanagawa, Kagoshima, and Fukuoka Prefectures, Japan (Table 1). Egg masses were obtained from more than 20 females for each regional population and maintained on an artificial diet (Insecta LFS or Silk Mate 2S, Nihon-Nosan Kogyo, Yokohama, Japan) at 25 °C under a 16:8 h light:dark photoperiod [20]. F2 or F3 generation larvae were used for bioassays for insecticide sensitivity and molecular diagnoses unless otherwise mentioned.
Male adults were collected using pheromone traps (SE trap, Sankei chemical, Kagoshima, Japan) equipped with pressure-sensitive adhesive sheets and synthetic sex pheromones of A. honmai (Shin-Etsu Chemical, Tokyo, Japan or Sumitomo Chemical, Tokyo, Japan). The traps were set on plucking surface of tea plants using agricultural poles in the field for 1–7 days.
Extraction of genomic DNA
For molecular diagnostics, abdomens of adults or larvae (4th–5th instar) were excised, individually placed into 0.2 ml PCR tubes containing 100 µl of an alkaline solution (50 mM NaOH, 0.2 mM EDTA), lightly crushed with a toothpick, and heated at 95 °C for 10 min. Supernatants (30 µl) were transferred to clean 0.2 ml PCR tubes containing 50 µl of 0.2 M Tris–HCl (pH 8.0), and mixed by pipetting. Extracts were diluted 100 times with distilled water (DW) and used as templates for PCR.
PCR-restriction fragment length polymorphism (PCR–RFLP)
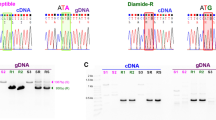
Previously, we have identified a candidate SNP marker for tebufenozide-resistance in A. honmai [18]. This SNP locates on exon 6 of the genomic DNA of EcR (Fig. 1a), which corresponds to C1244T in the EcR B1 isoform cDNA (Accession Nos. LC209228 for resistant type and LC209229 for susceptible type) and causes A415V substitution in the ligand binding domain of EcR protein in tebufenozide resistant strains. The genomic DNA region encompassing the A415V mutation of EcR (Fig. 1a) was amplified by PCR using primers F (5′-TGACGCTATTGTATTGTGGTTTC-3′) and R (5′-CTCAATGACGTAGGCCATG-3′). PCRs were performed in a reaction mixture containing 2.5 µl of EmeraldAmp MAX PCR Master Mix (TaKaRa Bio Inc., Kusatsu, Japan), 1 µl of template DNA, 0.13 µl of each primer (10 µM), and 1.3 µl of DW. PCR amplification was performed using an Applied Biosystems 2720 thermal cycler under following conditions: 94 °C for 30 s; followed by 40 cycles of 98 °C for 10 s, 65 °C for 30 s, and 72 °C for 30 s. Subsequently, 4.8 µl of DW and 0.2 µl of HhaI (New England Biolabs Japan, Tokyo, Japan) were added directly to the reactions and incubated at 37 °C for 1 h. Digested DNA fragments were separated by electrophoresis on a 2% agarose gel.
Schematics of PCR–RFLP genotyping of EcR A415V in Adoxophyes honmai. a Genomic DNA of A. honmai EcR was amplified by PCR using the designated primers, then digested with HhaI, which recognizes only the susceptible-specific allele (GCGC). b Electrophoretic analysis (2% agarose gel) of the PCR–RFLP products. Two bands (202 and 99 bp) were detected in susceptible homozygotes (S/S), one band (301 bp) was detected in resistant homozygotes (R/R), and three bands were detected in heterozygotes (R/S)
Rate of resistant (R)-allele (1244T or 415V) in a particular population was calculated as follows: R-allele rate (%) = 100 × [(R/R) × 2 + (R/S) × 1]/[((R/R) + (R/S) + (S/S)) × 2], where (R/R), (R/S), and (S/S) indicate the number of resistant homozygotes, resistant/susceptible heterozygotes, and susceptible homozygotes, respectively.
Direct sequencing of EcR genes
The EcR gene containing the A415V region was amplified from the genomic DNA by PCR using the F and R primers. PCR amplification was performed in a reaction mixture containing 6.25 µl of 2 × PCR Buffer for KOD FX Neo (TOYOBO, Osaka, Japan), 1 µl of template DNA, 2.5 µl of 2 mM dNTP mix, 0.37 µl of each primer (10 µM), 0.25 µl of KOD FX Neo polymerase, and 2.25 µl of DW. PCR amplification was performed using an Applied Biosystems 2720 thermal cycler under following conditions: 94 °C for 2 min, followed by 40 cycles of 98 °C for 10 s, 65 °C for 30 s, and 68 °C for 30 s. After PCR, 3 μl of the reaction mixture was mixed with 2 μl of Illustra ExoProStar (GE Healthcare, Tokyo, Japan) and heated at 37 °C for 15 min followed by 80 °C for 15 min, to prepare template DNA ready for direct-sequencing. Sequencing was performed in a reaction mixture containing 5 μl of template DNA, 2 μl of 5 × Sequencing Buffer, 1.2 μl of R primer (3 μM), and 1 μl of BigDye Terminator v3.1 Ready Reaction Mix (Thermo Fisher Scientific, Tokyo, Japan) 0.8 μl of DW. The conditions for the sequencing reaction were as follows: 96 °C for 1 min, followed by 25 cycles of 96 °C for 10 s, 55 °C for 5 s, and 60 °C for 4 min. DNA sequencing was performed using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Tokyo, Japan).
Bioassay for tebufenozide resistance
Tebufenozide resistance in A. honmai populations was evaluated using a leaf-dipping bioassay as described by Uchiyama and Ozawa [20]. Briefly, tea leaves were dipped with tebufenozide (Nihon Nohyaku, Tokyo, Japan), which was diluted 1000-fold using tap water with 0.01% Tween 20. Control leaves were treated with tap water containing only 0.01% Tween 20. Ten 2nd or 3rd instar larvae were placed on six treated leaves in a plastic dish and mortality was examined after 10 days. Each assay was performed in triplicate. Corrected mortality (%) was calculated using Abbott’s formula [1] as follows: 100 × (survival rate in the control − survival rate in the test)/survival rate in the control.
Statistical analysis
The Spearmann’s rank correlation coefficient between the corrected mortality and the frequencies of resistant allele was calculated using GraphPad Prism version 4.0 (GraphPad Software, Inc., California, USA). The regression equation between the corrected mortality and the R-allele rate was estimated based on a generalized nonlinear model. A binomial distribution was assumed for the probability distribution, and the survival probability (p) for a given R-allele rate (x) was assumed to be p = (a + b x)(1 − C), where C indicates the background mortality,and a and b are constants. Then, the corrected mortality (m) is given by m = 1 − (a + b x). The “bnlr” function of gnlm package in R [16] was used for the calculation. The actual R-script is available in supplementary information.
Results
PCR–RFLP method detecting A415V mutation in EcR
Genomic DNA extracted form A. honmai larvae was amplified by PCR using the primers F and R, and the amplicon was digested with HhaI (Fig. 1a). After an agarose gel electrophoresis of the digested products, two DNA bands (202 and 99 bp) were detected in susceptible homozygotes (S/S), while a single DNA band (301 bp) was detected in resistant homozygotes (R/R) (Fig. 1b). Three bands (301, 202, and 99 bp) were detected in resistant/susceptible heterozygotes (R/S) (Fig. 1b). Genotypes determined by the PCR–RFLP method on 8 larvae from 4 field populations (N = 8; Nos. 6, 13, 16, and 19 in Table 1) were in complete agreement with the results of DNA sequencing (Table 2), confirming that the PCR–RFLP method is reliable.
Genetic diagnosis of A. honmai regional populations
The A415V genotypes were determined by the PCR–RFLP method for 26 A. honmai populations collected from various tea fields in Japan (Table 1 and Fig. 2a). As a result, the R-allele was found in all populations collected in Shizuoka Prefecture (Nos. 1–11) (Fig. 2b). However, the frequencies of R-homozygotes varied depending on the population (i.e., ˃ 70% in Nos. 1–7, and ˂ 30% in Nos. 8–11). High frequencies (> 50%) of R-homozygotes were also found in two populations from Mie Prefecture (Nos. 12 and 13). The R-allele was also found in three out of four populations from Kyoto Prefecture (Nos. 14–16). However, the frequencies of R-homozygotes were relatively low (< 30%). No R-allele was found in the Kyoto population (No. 17), or in populations from Saitama, Ibaraki, Kanagawa, Kagoshima, and Fukuoka Prefectures (Nos. 18–26).
Map of collection sites of Adoxophyes honmai in the tea fields in Japan and genotyped rates of EcR mutation. a Insect collecting was performed in tea production areas of Shizuoka (Nos. 1–11), Mie (Nos. 12, 13), Kyoto (Nos. 14–17), Saitama (No. 18), Ibaraki (No. 19), Kanagawa (Nos. 20, 21), Kagoshima (Nos. 22–24), and Fukuoka (Nos. 25, 26) Prefectures in 2015 and 2016. Collection numbers have been designated in order of the highest proportion of resistant-allele rate. Numbers on the plot correspond to site collection numbers in Table 1. b Genotyped rates of EcR mutation. Genotypes were identified by PCR–RFLP analysis. Numbers on the plot correspond to site collection numbers in Table 1
Relationship between the A415V mutation and the magnitude of tebufenozide resistance
Figure 3 shows the relationship between the corrected mortality of larvae fed on leaves dipped with tebufenozide and the rate of R-allele. There is a strong correlation between the rate of R-allele and the corrected mortality (Spearman’s rank correlation coefficient r = − 0.8970, p < 0.0001, n = 24). This result supports our hypothesis that the A415V mutation of EcR is the principal factor for tebufenozide resistance. Furthermore, the regression analysis based on the generalized nonlinear model showed regression equation of y = 99.45 − 0.89 x, where y is corrected mortality (%) and x is R-allele rate. Using this equation, the magnitude of tebufenozide resistance (corrected mortality) can be accurately estimated by the R-allele rate.
Correlation of resistant-allele rate with corrected mortality owing to tebufenozide. Numbers on the plot correspond to site collection numbers in Table 1. Corrected mortalities of Nos. 1, 4–6, 12–16, 18, 19, and 22 were the same as reported previously [18]. Twenty-eight to thirty F2 or F3 larvae were analyzed for each population. The dotted line indicates regression equation (y = 99.45 − 0.89 x) calculated based on the generalized linear model
Genotyping of adults collected by pheromone traps
We applied the PCR–RFLP method on adult males collected by pheromone traps. Consequently, 3361 out of 3436 pheromone-trapped adults were successfully genotyped. The detection rate of adults (3361/3436; 97.8%) was comparable to that of larvae (591/597; 99.0%). In addition, we compared the rates of R-alleles determined by PCR–RFLP using pheromone-trapped adults and laboratory-reared larvae (F2) collected from same locations. As shown in Table 3, similar values were obtained for adults and larvae in all local populations examined.
Discussion
In this study, we developed a molecular diagnostic test for tebufenozide resistance in A. honmai—a PCR–RFLP method to detect the A415V mutation of EcR. Using the PCR–RFLP method, a high correlation was found between the R-allele rate and the magnitude of tebufenozide resistance in various regional populations. This result further confirmed the conclusion of our previous study that the A415V mutation of EcR is a major factor in resistance to tebufenozide [18].
The PCR–RFLP method was successfully applied not only to laboratory-reared larvae, but also to pheromone-trapped adults (Table 3). This result indicates that the PCR–RFLP method may be used as a reliable and convenient tool for field-monitoring of tebufenozide-resistance.
The detection rate of pheromone-trapped adults (97.8%) was as high as that of laboratory-reared larvae (99.0%). It is a bit surprising because the former was expected to be decreased by DNA degradation owing to environmental factors (i.e., UV-radiation, high humidity, and high temperature). In the diamondback moths, P. xylostella, UV-radiation accelerates DNA degradation in pheromone-trapped adults in the summer, where changing traps weekly is preferable to maintain a detection rate ˃ 90% [22]. In contrast, we observed high detection rates (96.0%; 24/25, data not shown) from pheromone-trapped samples left for 1 month in the field. Therefore, the sampling duration for A. honmai could be longer than for P. xylostella. However, we recommend changing traps every 1–2 weeks or 2–3 days when population densities are high, to avoid exceeding the maximum capture number per trap (800–900). Biological factors (i.e., fungi, bacteria, predators, etc.) can also deteriorate samples and affect detection rate, as we often observed severe feeding damage of trapped samples by snails. Pasting a repellent agent on the trap edge is also recommended to protect from snail predation.
Molecular diagnostics suggest that the selection of the R-allele was critical for the development of tebufenozide resistance in A. honmai in the Shizuoka Prefecture. Populations collected from Shimada (No. 1), Makinohara (Nos. 4 and 6), and Kikugawa city (No. 5)—located in the Makinohara Plateau area—showed high tebufenozide resistance, while populations collected from Kakegawa (No. 8), Omaezaki (No. 9), and Shizuoka city (No. 11)—located outside of the Makinohara Plateau area—showed low to moderate resistance. R-homozygotes were dominant (> 80%) in the Makinohara Plateau areas, and less dominant (< 30%) in the other areas. Tea plants are widely cultivated in the areas located outside the Makinohara Plateau areas, while open-field and greenhouse vegetables (i.e., strawberry and melon) are cultivated in addition to tea plants. We speculate that the more extensive and continuous exposure to tebufenozide used for the protection against A. honmai in the Makinohara Plateau accelerated the selection of the R-allele, and thus the development of tebufenozide-resistance.
Field populations showing moderate to high tebufenozide resistance were found in two (Nos. 12 and 13) and three (Nos. 14, 15, and 16) areas in Mie and Kyoto Prefectures, respectively (Fig. 3b). This is the first report of tebufenozide resistance in A. honmai outside of Shizuoka Prefecture [20]. R-homozygotes were also found in these populations, suggesting that the mechanism of tebufenozide resistance is the same as the populations in the Shizuoka Prefecture. However, it remains unclear whether the A415V mutation of EcR developed independently of populations in the Shizuoka Prefecture and were retained in local populations through positive selection.
Migration is a critical factor for the dispersal rate of resistance. Plutella xylostella has the ability to fly over 3,000 km [17]; therefore, insecticide resistance in this species is rapidly spread over wide areas via migratory activity. For example, diamide resistance in P. xylostella was first discovered in Kagoshima Prefecture in 2013 [4], and rapidly spread to Hokkaido Prefecture in 2014. By contrast, tebufenozide resistance in A. honmai was first reported in Shizuoka Prefecture 1997 [7] and was not detected in other Prefectures for 21 years. A flight mill analysis showed that A. honmai were only capable of flying approx. 5 km per night [14], suggesting the lower flight ability as a factor in the slow expansion of tebufenozide resistance. Future investigations will reveal the actual extent of migration in A. honmai in Japan using molecular markers such as EcR, cytochrome oxidase I [8], and other genes.
In conclusion, this study reported a diagnostic method that targets the A415V mutation of EcR in A. honmai. This method would be a promising tool for monitoring tebufenozide resistance in the field and contribute to the development of management systems for this destructive plant pest.
References
Abbott, W. S. (1925). A method of computing the effectiveness of an insecticide. Journal of Economic Entomology, 18, 265–267.
Cao, G., & Han, Z. (2006). Tebufenozide resistance selected in Plutella xylostella and its cross-resistance and fitness cost. Pest Management Science, 62, 746–751.
Dunley, J. E., Brunner, J. F., Doerr, M. D., & Beers, E. H. (2006). Resistance and cross-resistance in populations of the leafrollers, Choristoneura rosaceana and Pandemis pyrusana, in Washington apples. Journal of Insect Science, 6, 1–7.
Fukuda, K., & Hayashikawa, S. (2014). Decline in susceptibility to diamide insecticides of the diamondback moth, Plutella xylostella L. (Lepidoptera), collected in Kagoshima Prefecture. Kyushu Pl Prot Res, 60, 75–78. (in Japanese).
Ioriatti, C., Tasin, M., Charmillot, P. J., Reyes, M., & Sauphanor, B. (2007). Early detection of resistance to tebufenozide in field populations of Cydia pomonella L.: Methods and mechanisms. Journal of Applied Entomology, 131, 453–459.
Jia, B., Liu, Y., Zhu, Y. C., Liu, X., Gao, C., & Shen, J. (2009). Inheritance, fitness cost and mechanism of resistance to tebufenozide in Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae). Pest Management Science, 65, 996–1002.
Kosugi, Y. (1999). Decline in the susceptibility of smaller tea tortrix, Adoxophyes honmai to some insecticides in Shimada city, Shizuoka Prefecture. Annual Report of the Kanto-Tosan Plant Protection Society, 46, 123–126. (in Japanese).
Lee, S. Y., Park, H., Boo, K. S., Park, K. T., & Cho, S. (2005). Molecular identification of Adoxophyes honmai (Yasuda) (Lepidoptera: Tortricidae) based on mitochondrial COI gene sequences. Molecules and Cells, 19, 391–397.
Minamikawa, J., & Osakabe, M. (1979). Insect pest of the tea plant (p. 322). Tokyo: Japan Plant Protection Association. (in Japanese).
Moulton, J. K., Pepper, D. A., Jansson, R. K., & Dennehy, T. J. (2002). Pro-active management of beet armyworm (Lepidoptera: Noctuidae) resistance to tebufenozide and methoxyfenozide: Baseline monitoring, risk assessment, and isolation of resistance. Journal of Economic Entomology, 95, 414–424.
Oberlander, H., Silhacek, D. L., & Porcheron, P. (1995). Non-steroidal ecdysteroid agonists: Tools for the study of hormonal action. Archives of Insect Biochemistry and Physiology, 28, 209–223.
Piyasaengthong, N., Sato, Y., Kinoshita, N., & Kainoh, Y. (2016). Oviposition preference for leaf age in the smaller tea tortrix, Adoxophyes honmai (Lepidoptera: Tortricidae) as related to performance of neonates. Applied Entomology and Zoology, 51, 363–371.
Shirai, M., Kobayashi, H., Ito, H., Hotta, K., & Takeshima, S. (1988). Decline in the susceptibility of Homona magnanima Diaknoff to methomyl in Shizuoka Prefecture. Annual Report of the Kanto-Tosan Plant Protection Society, 35, 189–190. (in Japanese).
Shirai, Y., & Kosugi, Y. (2000). Flight activity of the smaller tea tortrix, Adoxophyes honmai (Lepidoptera: Tortricidae). Applied Entomology and Zoology, 35, 459–466.
Smirle, M. J., Lowery, D. T., & Zurowski, C. L. (2002). Resistance and cross-resistance to four insecticides in populations of obliquebanded leafroller (Lepidoptera: Tortricidae). Journal of Economic Entomology, 95, 820–825.
Swihart, L. (2017). gnlm: Generalized nonlinear regression models. R package version 1.1.0. https://CRAN.R-project.org/package=gnlm. Accessed 03 Feb 2019.
Talekar, N. S., & Shelton, A. M. (1993). Biology, ecology, and management of the diamondback moth. Annual Review of Entomology, 38, 275–301.
Uchibori-Asano, M., Jouraku, A., Uchiyama, T., Yokoi, K., Akiduki, G., Suetsugu, Y., et al. (2019). Genome-wide identification of Tebufenozide resistant genes in the smaller tea tortrix, Adoxophyes honmai (Lepidoptera: Tortricidae). Scientific Reports. https://doi.org/10.1038/s41598-019-40863-5.
Uchiyama, T., Ozawa, A., & Yoo, J. (2013). Susceptibility and resistance to diacylhydrazine analog insect growth regulator insecticides in the smaller tea tortrix, Adoxophyes honmai Yasuda (Lepidoptera: Tortricidae), collected in tea fields in Shizuoka Prefecture, Japan. Journal of Applied Entomology and Zoology, 57, 85–93. (in Japanese with English summary).
Uchiyama, T., & Ozawa, A. (2014). Rapid development of resistance to diamide insecticides in the smaller tea tortrix, Adoxophyes honmai (Lepidoptera: Tortricidae), in the tea fields of Shizuoka Prefecture, Japan. Applied Entomology and Zoology, 49, 529–534.
Uchiyama, T., & Ozawa, A. (2015). Inheritance of tebufenozide resistance in the smaller tea tortrix, Adoxophyes honmai (Lepidoptera: Tortricidae). Journal of Applied Entomology and Zoology, 59, 127–131. (in Japanese with English summary).
Uesugi, R., Hinomoto, N., & Goto, C. (2016). Estimated time frame for successful PCR analysis of diamondback moths, Plutella xylostella (Lepidoptera: Plutellidae), collected from sticky traps in field conditions. Applied Entomology and Zoology, 51, 505–510.
Wearing, C. H. (1998). Cross-resistance between azinphos-methyl and tebufenozide in the greenheaded leafroller, Planotortrix octo. Pesticide Science, 54, 203–211.
Wing, K. D. (1988). RH 5849, a nonsteroidal ecdysone agonist: Effects on a Drosophila cell line. Science, 241, 467–469.
Wing, K. D., Slawecki, R. A., & Carlson, G. R. (1988). RH 5849, a nonsteroidal ecdysone agonist: Effects on larval Lepidoptera. Science, 241, 470–472.
Acknowledgements
We are grateful to Chiharu Tanaka of the Mie Prefecture Agricultural Research Institute; Koji Yamashita of the Kyoto Prefectural Agriculture, Forestry and Fisheries Technology Center; Takeshi Iwasaki of Saitama Tea Research Institute; Tomoki Sekizuka and Shigeru Watanabe of Kanagawa Agricultural Technology Center; Takeshi Kamimuro of the Kagoshima Prefectural Institute for Agricultural Development, and Tetsuya Yoshioka and Rie Inoue of the Fukuoka Agriculture and Forestry Research Center for providing insect samples. We thank Utako Takano and Akemi Inoue in NARO and Junko Sumikawa, Tazuko Onikubo and Rie Takimoto of the Shizuoka Research Institute of Agriculture and Forestry for assistance with insect rearing and DNA extraction. This research was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics-based Technology for Agricultural Improvement; PRM-02).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Uchibori-Asano, M., Uchiyama, T., Jouraku, A. et al. Tebufenozide resistance in the smaller tea tortrix, Adoxophyes honmai (Lepidoptera: Tortricidae): establishment of a molecular diagnostic method based on EcR mutation and its application for field-monitoring. Appl Entomol Zool 54, 223–230 (2019). https://doi.org/10.1007/s13355-019-00616-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13355-019-00616-2