Abstract
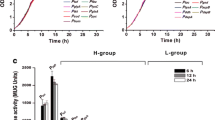
The replacement of promoters with various strengths is an effective strategy to fine-tune gene expression. More available promoters with a broad range of transcription efficiency are needed in metabolic engineering. In this study, a putative protein coding gene CP_2454 was identified with a stable and high transcriptional level from an l-leucine-producing strain Corynebacterium glutamicum CP, which was absent in wild-type C. glutamicum ATCC 13032. The transcriptional level of CP_2454 was about 80.0% those of tuf and sod, and was 1.6 and 3.2 times those of ilvB and gapA, respectively. These promoters were cloned into C. glutamicum ATCC 13032 to control the expression of green fluorescent protein (GFP). While, the expressional level of GFP under the control of PCP_2454 was close to that of Ptuf and Psod, which was significantly higher than that of other tested promoters. The native promoter of ilvB controlling the expression of a feedback-resistant acetolactate synthase was replaced by PCP_2454, resulting in an increase of l-valine titer by 58.5%. A further 24.9% increase of l-valine titer was achieved after replacement of the promoter of ilvD encoding dihydroxyacid dehydratase with PCP_2454. Identification of the constitutive promoter PCP_2454 expands the promoter library of C. glutamicum and provides essential reference for tunable expression of target genes in C. glutamicum.




Similar content being viewed by others
References
Akyol I, Comlekcioglu U, Karakas A, Serdaroglu K, Ekinci MS, Ozkose E (2008) Regulation of the acid inducible rcfB promoter in Lactococcus lactis subsp. lactis. Ann Microbiol 58:269–273
Baumgart M, Frunzke J (2015) The manganese-responsive regulator MntR represses transcription of a predicted ZIP family metal ion transporter in Corynebacterium glutamicum. FEMS Microbiol Lett 362:1–10
Chung SC, Park JS, Yun J (2017) Improvement of succinate production by release of end-product inhibition in Corynebacterium glutamicum. Metab Eng 40:157–164
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12:499–510
Gerstmeir R, Wendisch VF, Schnicke S, Ruan H, Farwick M, Reinscheid D, Eikmanns BJ (2003) Acetate metabolism and its regulation in Corynebacterium glutamicum. J Biotechnol 104:99–122
Gui Y, Ma Y, Xu Q, Zhang C, Xie X, Chen N (2016) Complete genome sequence of Corynebacterium glutamicum CP, a Chinese l-leucine producing strain. J Biotechnol 220:64–65
Harnpicharnchai P, Promdonkoy P, Sae-Tang K, Roongsawang N, Tanapongpipat S (2014) Use of the glyceraldehyde-3-phosphate dehydrogenase promoter from a thermotolerant yeast, Pichia thermomethanolica, for heterologous gene expression, especially at elevated temperature. Ann Microbiol 64:1457–1462
Ikeda M, Mizuno Y, Awane S, Hayashi M, Mitsuhashi S, Takeno S (2011) Identification and application of a different glucose uptake system that functions as an alternative to the phosphotransferase system in Corynebacterium glutamicum. Appl Microbiol Biotechnol 90:1443–1451
Jakoby M, Ngouoto-Nkili CE, Burkovski A (1999) Construction and application of new Corynebacterium glutamicum vectors. Biotechnol Tech 13:437–441
Kind S, Neubauer S, Becker J, Yamamoto M, Völkert M, Abendroth GV, Zelder O, Wittmann C (2014) From zero to hero—production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum. Metab Eng 25:113–123
Liu Y, Zhuang Y, Ding D, Xu Y, Sun J, Zhang D (2017) Biosensor-based evolution and elucidation of a biosynthetic pathway in Escherichia coli. ACS Synth Biol 6:837–848
Malakar P, Venkatesh KV (2012) Effect of substrate and IPTG concentrations on the burden to growth of Escherichia coli on glycerol due to the expression of Lac proteins. Appl Microbiol Biotechnol 93:2543–2549
Nešvera J, Pátek M (2011) Tools for genetic manipulations in Corynebacterium glutamicum and their applications. Appl Microbiol Biotechnol 90:1641–1654
Neuner A, Heinzle E (2015) Mixed glucose and lactate uptake by Corynebacterium glutamicum through metabolic engineering. Biotechnol J 6:318–329
Okibe N, Suzuki N, Inui M, Yukawa H (2010) Isolation, evaluation and use of two strong, carbon source-inducible promoters from Corynebacterium glutamicum. Lett Appl Microbiol 50:173–180
Pátek M, Nešvera J (2011) Sigma factors and promoters in Corynebacterium glutamicum. J Biotechnol 154:101–113
Plassmeier JK, Busche T, Molck S, Persicke M, Pühler A, Rückert C, Kalinowski J (2013) A propionate-inducible expression system based on the Corynebacterium glutamicum prpD2, promoter and PrpR activator and its application for the redirection of amino acid biosynthesis pathways. J Biotechnol 163:225–232
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73
Scheele S, Oertel D, Bongaerts J, Evers S, Hellmuth H, Maurer KH, Bott M, Freudl R (2013) Secretory production of an FAD cofactor-containing cytosolic enzyme (sorbitol–xylitol oxidase from Streptomyces coelicolor) using the twin-arginine translocation (Tat) pathway of Corynebacterium glutamicum. Microb Biotechnol 6:202–206
Siebert D, Wendisch VF (2015) Metabolic pathway engineering for production of 1,2-propanediol and 1-propanol by Corynebacterium glutamicum. Biotechnol Biofuels 8:91
Solovyev V, Salamov A (2011) Automatic annotation of microbial genomes and metagenomic sequences. Metagenomics Appl Agric Biomed Environ 2011:61–78
Sun Y, Guo W, Wang F, Peng F, Yang Y, Dai X, Liu X, Bai Z (2016) Transcriptome and multivariable data analysis of Corynebacterium glutamicum under different dissolved oxygen conditions in bioreactors. PLoS One 11:e0167156
Tateno T, Fukuda H, Kondo A (2007) Direct production of l-lysine from raw corn starch by Corynebacterium glutamicum secreting Streptococcus bovis alpha-amylase using cspB promoter and signal sequence. Appl Microbiol Biotechnol 77:533–541
Teramoto H, Inui M, Yukawa H (2011) Transcriptional regulators of multiple genes involved in carbon metabolism in Corynebacterium glutamicum. J Biotechnol 154:114–125
Toyoda K, Inui M (2016) Regulons of global transcription factors in Corynebacterium glutamicum. Appl Microbiol Biotechnol 100:45–60
Vogt M, Haas S, Klaffl S, Polen T, Eggeling L, van Ooyen J, Bott M (2013) Pushing product formation to its limit: metabolic engineering of Corynebacterium glutamicum for l-leucine overproduction. Metab Eng 22:40–52
Yim SS, An SJ, Kang M, Lee J, Jeong KJ (2013) Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol Bioeng 110:2959–2969
Funding
This study was supported by the National Natural Science Foundation of China (31470211 and 31770053), Natural Science Foundation of Tianjin (17JCQNJC09500), and Tianjin Municipal Science and Technology Commission (17YFZCSY01050).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
ESM 1
(PDF 69.6 kb)
Rights and permissions
About this article
Cite this article
Wei, H., Ma, Y., Chen, Q. et al. Identification and application of a novel strong constitutive promoter in Corynebacterium glutamicum. Ann Microbiol 68, 375–382 (2018). https://doi.org/10.1007/s13213-018-1344-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-018-1344-0