Abstract
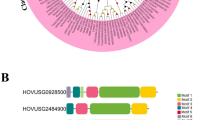
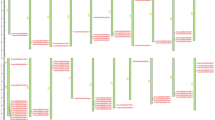
Plants have evolved a series of mechanisms to resist pathogens infection. The nucleotide-binding site and leucine-rich repeat (NBS-LRR) family contains the largest number of plant disease resistance genes in plants. Recognition of Peronospora Parasitica 13-like (RPP13-like) genes belong to this superfamily and play important roles in the resistance of various plant diseases including the downy mildew caused by Peronospora parasitica. In this study, 21 RPP13-like genes were identified in barley via bioinformatics. These genes all contained CC, NB-ARC and LRR domains. The physical and chemical properties, chromosome locations, gene structures, protein motifs, 3D protein structures, and microarray based expression dynamics of these genes, as well as their phylogenetic relationship with other plant species were analyzed. Non-expression of MLOC_19262.1 was detected without pathogen infection. When barley was inoculated with the powdery mildew pathogenic fungus, the expression of MLOC_ 19262.1 reached a very high level, suggesting that this gene is an important and promising candidate resistance gene for further study. The two RPP13-like genes, MLOC_57007.2 and MLOC_5059.1 may be involved in barley regular or abiotic stress induced physiological metabolism in specific tissues or at specific developmental stages; furthermore, these functions may be associated with specific domains. These findings provided evidence for the functional diversity of plant pathogen resistance genes and will be helpful for the future characterization of the PRR13-like gene subfamily.
Similar content being viewed by others
References
Jones, J.D. & Dang, I.J. The plant immune system. Nature 444, 323–329 (2006).
Li, B., Meng, X.Z., Shan, L.B. & He, P. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host & Microbe. 19, 641–650 (2016).
Allen, R.L. et al. Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306, 1957–1960 (2004).
Dodds, P.N. & Schwechheimer, C.A. Breakdown in defense signaling. Plant Cell 14, 5–8 (2002).
Wang, X.S., Wu, W.R., Jin, G.L. & Zhu, J. Genomewide identification of R genes and exploitation of candidate RGA markers in rice. Chinese Sci. Bull. 50, 1120–1125 (2005).
Dodds, P.N., Lawrence, G.J. & Ellis, J.G. Six amino acid changes confined to the leucine-rich repeat β-Strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13, 163–178 (2001).
Crute, L.R. & Pink, D.A. Genetics and utilization of pathogen resistance in plants. Plant Cell 8, 1747–1755 (1996).
Bittner-eddy, P.D., Crute, L.R., Holub, E.B. & Beynon, J.L. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 21, 177–188 (2000).
Boyes, D.C., Nam, J. & Dangl, J.L. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. PNAS 95, 15849–15854 (1998).
Rose, L.E. et al. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics. 166, 1517–1527 (2004).
Jia, Y.L., McAdams, S.A., Bryan, G.T., Hershey, H.P. & Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014 (2000).
Amin, N., Ali, B., Muzaffar, S., Majeed, I. & Sayeed, N. Symptoms, pathogen biology and control of downy mildew of brassica. J. Biosphere. 2, 6–9 (2013).
Bittner-Eddy, P.D. et al. Genetic and physical mapping of the RPP13 locus, in Arabidopsis, responsible for specific recognition of several Peronospora parasitica (downy mildew) isolates. Mol. Plant Microbe. In. 12, 792–802 (1999).
Liang, Y. et al. Genome-wide identifcation, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus. Sci. Rep. 6, 24265 (2016).
Martin, G.B., Bogdanove, A.J. & Sessa, G. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant. Biol. 54, 23–61 (2003).
Bittner-eddy, P.D. & Beynon, J.L. The Arabidopsis downy mildew resistance gene, RPP13-Nd, functions independently of NDR1 and EDS1 and does not require the accumulation of salicylic acid. Mol. Plant Microbe. In. 14, 416–421 (2001).
Wan, H. et al. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genomics 14, 109 (2013).
Pandey, S.P. & Somssich, I.E. The Role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655 (2009).
Song, X.M. et al. Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica ra-passp. pekinensis). Genomics 14, 135–146 (2014).
Caldo, R.A., Nettleton, D., Peng, J. & Wise, R.P. Stage-specific suppression of basal defense discriminates barley plants containing fast and delayed-acting Mla powdery mildew resistance alleles. Mol. Plant Microbe. In. 19, 939–947 (2006).
Hruz, T. et al. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 420747, 35–39 (2008).
Muñoz-Amatriaín, M. et al. Transcriptome analysis of a barley breeding program examines gene expression diversity and reveals target genes for malting quality improvement. BMC Genomics 11, 1–15 (2010).
Abebe, T., Melmaiee, K., Berg, V. & Wise, R.P. Drought response in the spikes of barley: gene expression in the lemma, palea, awn, and seed. Func. Integr. Genomic. 10, 191–205 (2010).
Gómez-Porras, J.L., Riaño-Pachón, D.M., Dreyer, I., Mayer, J.E. & Mueller-Roeber, B. Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics 8, 260 (2007).
Goldsbrough, A.P., Albrecht, H. & Stratford, R. Salicylic acid inducible binding of a tobacco nuclear protein to a 10 bp sequence which is highly conserved amongst stress-inducible genes. Plant J. 3, 563–571 (1993).
Tan, C.M. et al. OsPOP5, a prolyl oligopeptidase family gene from rice confers abiotic stress tolerance in Escherichia coli. Int. J. Mol. Sci. 14, 20204–20219 (2013).
Zhang, X.L., Yang, Z.P., Zhang, J. & Zhang, L.G. Ectopic expression of BraYAB1-702, a member of YABBY gene family in Chinese cabbage, causes leaf curling, inhibition of development of shoot apical meristem and flowering stage delaying in Arabidopsis thaliana. Int. J. Mol. Sci. 14, 14872–14891 (2013).
Pellegrineschi, A. et al. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 47, 493–500 (2004).
Bent, A.F. Plant disease resistance genes: function meets structure. The Plant Cell 8, 1757–1771 (1996).
Brown, N.J. et al. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331, 1436–1439 (2011).
Wilkinson, S. & Davies, W.J. Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant, Cell & Environ. 33, 510–525 (2010).
Umezawa, T. et al. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839 (2010).
Nakashima, K. et al. Transcriptional regulation of ABI3-and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant. Mol. Biol. 60, 51–68 (2006).
Hobo, T., Asada, M., Kowyama, Y. & Hottori, T. ACGTcontaining abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19, 679–689 (1999).
Yanagisawa, S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 21, 281–288 (2000).
Rejeb, I.B., Pastor, V. & Mauch-Mani, B. Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants 3, 458–475 (2014).
Ramegowda, V. & Senthil-Kumar, M. The interactive effects of simultaneous biotic and abiotic stresses on plants: mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 176, 47–54 (2015).
Rojas, C.M., Senthil-Kumar, M., Tzin, V. & Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 5, 1–12 (2014).
Goodstein, D.M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, 1178–1186 (2012).
Gasteiger, E. et al. Protein identification and analysis tools in the ExPASy server. Method. Mol. Biology 112, 531–552 (1999).
Lin, Y.X. et al. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12, 1–14 (2011).
Bailey, T.L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, w202–w208 (2009).
Källberg, M. et al. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 (2012).
Lovell, S.C. et al. Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins: Structure Function & Bioinformatics 50, 437–450 (2003).
Deng, W., Wang, Y., Liu, Z., Cheng, H. & Xue, Y. HemI: a toolkit for illustrating heatmaps. Plos One 9, e111988–e111988 (2014).
Lescot, M. et al. PlantCARE: a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327 (2002).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Cheng, J., Fan, H., Li, L. et al. Genome-wide Identification and Expression Analyses of RPP13-like Genes in Barley. BioChip J 12, 102–113 (2018). https://doi.org/10.1007/s13206-017-2203-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-017-2203-y