Abstract
Separate murine knockout (KO) of either c- or N-myc genes in neural stem and precursor cells (NSC) driven by nestin-cre causes microcephaly. The cerebellum is particularly affected in the N-myc KO, leading to a strong reduction in cerebellar granule neural progenitors (CGNP) and mature granule neurons. In humans, mutation of N-myc also causes microcephaly in Feingold Syndrome. We created a double KO (DKO) of c- and N-myc using nestin-cre, which strongly impairs brain growth, particularly that of the cerebellum. Granule neurons were almost absent from the Myc DKO cerebellum, and other cell types were relatively overrepresented, including astroglia, oligodendrocytes, and Purkinje neurons. These findings are indicative of a profound disruption of cell fate of cerebellar stem and precursors. DKO Purkinje neurons were strikingly lacking in normal arborization. Inhibitory neurons were ectopic and exhibited very abnormal GAD67 staining patterns. Also consistent with altered cell fate, the adult DKO cerebellum still retained a residual external germinal layer (EGL). CGNP in the DKO EGL were almost uniformly NeuN and p27KIP1 positive as well as negative for Math1 and BrdU at the peak of normal cerebellar proliferation at P6. The presence of some mitotic CGNP in the absence of S phase cells suggests a possible arrest in M phase. CGNP and NSC metabolism also was affected by loss of Myc as DKO cells exhibited weak nucleolin staining. Together these findings indicate that c- and N-Myc direct cerebellar development by maintaining CGNP and NSC populations through inhibiting differentiation as well as directing rapid cell cycling and active cellular metabolism.
Similar content being viewed by others
Introduction
Myc is most well known for its role in tumorigenesis when overexpressed including medulloblastomagenesis [1–6], but at physiological levels myc genes are important regulators of many aspects of normal cellular biological behavior including cellular metabolism and cell cycling (Reviewed in [7]). Myc genes encode atypical members of the basic-helix-loop-helix zipper transcription factor superfamily, which can activate and repress the transcription of specific genes. In addition in some cells including cerebellar granule neural progenitors (CGNP), they can act more globally on chromatin to maintain euchromatic domains associated with specific histone modifications such as acetylation of lysine 9 and methylation of lysine 4 of histone H3, as well as others including modifications of histone H4 [8–13].
While the vast majority of Myc studies have been conducted in tumor cells, there is growing evidence of key roles for myc genes at endogenous levels in both somatic and embryonic stem cells. Constitutive knockout (KO) of c- or N-myc causes embryonic lethality around midgestation [14, 15]. Conditional disruption of N-myc in neural stem and progenitor cells (NSC) severely disrupts murine brain growth, particularly that of the cerebellum, while a similar KO of c-myc moderately impairs growth [16, 17]. Disruption of either c- or N-myc or both in hematopoietic stem cells (HSC) also alters their normal biological functions, affecting survival and self-renewal [18, 19]. These studies support the hypothesis that every normal cell has a requirement for some minimum level of myc. myc genes also are involved in the production of induced pluripotent stem (iPS) cells (reviewed in [20]) [21–25]. While exogenous Myc is not formally required for the process [26, 27], it dramatically enhances the efficiency and in its absence its function is likely supplanted by endogenous myc. Data suggests that during iPS cell formation, Myc represses differentiation-associated genes [28] and may not have a key role in directly maintaining expression of pluripotency factors. However, in neuroblastoma and other tumors, some pluripotency genes such as lif, lin28b, Klf2, and Klf4 are N-Myc targets for activation, while a subset of these genes is also regulated in NSC by N-Myc [29].
Perhaps because of the importance of myc genes in normal cellular biology and their ability to cause cancer when in excess, cells have evolved systems to maintain normal total cumulative myc RNA and Myc protein levels. These include cross-regulation, redundancy and compensation between the three main myc genes—c-, N-, and L-myc—as well as Myc-trigged apoptosis when in excess. Myc protein stability is also tightly controlled (reviewed in [30]). While desirable for the cell and organism, this interconnectedness has made it more difficult for the biologist to discern the specific roles of individual myc genes through methods such as KO studies. For example, despite fairly ubiquitous expression in the developing brain and some other regions of the embryo, L-myc constitutive KO was reported to have no phenotype at all [31]. One theory is that this lack of apparent phenotype was due to the continued presence of N-myc and perhaps c-myc, which could fulfill the roles of L-myc. In contrast, constitutive KO of either c- or N-myc resulted in midgestational lethality [14, 15, 32], suggesting their overall embryonic functions are to some critical degree unique or their loss cannot be compensated for. Conditional double knockout (DKO) of c- and N-myc in hematopoietic stem cells yields a far more severe phenotype than disruption of either gene alone, suggesting additive or redundant roles [19]. A large degree of redundancy is also supported by the knocking of N-myc into the c-myc locus largely rescuing the loss of c-myc [33]. The prevailing theory is that what is most critical is the total level of all myc gene expression in each cell.
During neurogenesis, N-myc plays a particularly important role in NSC to direct overall brain and particularly cerebellar growth [16, 34], consistent with its fairly widespread expression pattern in the brain throughout embryogenesis including very high levels in the developing cerebellum. L-myc is also fairly widely expressed, particularly in the early midbrain, which suggests it may function in the brain despite no reported KO phenotype. Although c-myc expression has been less clearly defined, it appears to be more restricted than that of L- and N-myc. In vitro overexpression and c-myc KO studies in rat neurospheres support a crucial role for myc genes overall in NSC function and suggested functional ties to p53/Arf [35], but the relevance for in vivo neurogenesis remained unclear. While no nestin-cre L-myc conditional knockout has been reported, single nestin-cre-driven knockout of c-myc in NSC impairs brain growth, but far less severely than loss of N-myc and with no cerebellar specific phenotype [16, 34]. The molecular basis of the c- and N-myc nestin-cre-driven brain phenotypes is not completely clear, but in the N-myc knockout changes in cyclin D2 and cyclin-dependent kinase inhibitors such as p18INK4C and p27KIP1 were evident suggesting cell cycle regulation plays an important role [16, 36], including in the cerebellum. However, no unbiased global analysis of gene expression has been conducted for myc genes using a loss of function model in the developing brain or specifically in the cerebellum. Upstream of N-myc are several potential signaling pathways including Shh in the cerebellum and in CGNPs [17, 37], while N-myc also appears to be a critical part of a newly defined pathway DLL3 and Notch signaling in which N-Myc protein is targeted by the Huwe1 ubiquitin ligase [38, 39].
To address the endogenous functions of c- and N-myc in cerebellar development, we created a nervous system specific double c- and N-myc KO mouse by crossing doubly homozygously myc floxed mice with nestin-cre, which is expressed specifically in NSC and at very high levels in CGNP and in NSC of the cerebellum neuroepithelium. The myc DKO mice have a nervous system phenotype much more severe than either single myc KO alone, which is ultimately lethal. In terms of mechanisms, we found evidence that Myc regulates transcriptional programs including that of metabolic genes essential for normal stem and precursor fate during cerebellar genesis.
Methods and Methods
Animals
Breeding pairs of nestin-Cre− N-myc (FL/FL) c-myc (FL/FL) and nestin-Cre+ N-myc (FL/WT) c-myc (FL/FL) [10] were set up to generate the four genotypes Nestin-Cre− N-myc (FL/FL) c-myc (FL/FL), Nestin-Cre− N-myc (FL/WT) c-myc (FL/FL), Nestin-Cre+ N-myc (FL/FL) c-myc (FL/FL), and nestin-Cre+ N-myc (FL/WT) c-myc (FL/FL). Genotypes of animals were determined via polymerase chain reaction (PCR) using primer sets for Cre, N-myc and c-myc. Primer sequences were as follow: Cre1: GCC TGC ATT ACC GGT CGA TGC AAC GA; Cre2: GTG GCA GAT GGC GCG GCA ACA ACC ATT; N-myc1: GTC GCG CTA GTA AGA GCT GAG ATC; N-myc2: GGC ACA CAC CTA TAA TCC CAG CTA; N-myc3: CAC AGC TCT GGA AGG TGG GAG AAA GTT GAG CGT CTC C; c-myc1: GCC CCT GAA TTG CTA GGA AGA CTG; c-myc2: CCG ACC GGG TCC GAG TCC CTA TT. Cre1 and 2 primer set amplified a 700 bp band from Cre insertion. The flox-specific band for N-myc1, 2, and 3 primers is 260 bp, the wild-type band is 217 bp, and the deletion band is 350 bp. The flox-specific band for c-myc1 and 2 primer set is 500 bp, and the wild-type band is 400 bp. A separate primer set to detect c-myc deletion, c-mycDS: TCG CGC CCC TGA ATT GCT AGG AA, and c-mycDA: TGC CCA GAT AGG GAG CTG TGA TAC TT, were used and amplifies a band at ~750 bp.
Immunohisto- and Cytochemistry
All E12.5 embryos were immersion-fixed overnight in fresh, buffered 4% paraformaldehyde, paraffin embedded and cut (12 μm) sagittally. Pregnant female were anesthetized with 150 mg/kg ketamine and 16 mg/kg xylazine, then E17.5 embryos were removed individually and perfused with buffered 4% paraformaldehyde. Brains were removed and fixed overnight in fresh, buffered 4% paraformaldehyde, then cryopreserved and cut (12 μm) sagittally. For immunostaining, E12.5 embryo sections were deparaffinized then rehydrated and 30 mM sodium citrate treated for 10 min at 95°C, while E17.5 frozen brain sections were post-fixed in −20°C Acetone for 10 min followed by three 5 min PBS wash. All sections were blocked in 10% normal goat serum in PBS for 1 h at room temperature then incubated in primary antibody overnight at 4°C. Antibodies used include anti-BrdU (Chemicon MAB3424, 1:150), anti-PhosphoH3 (Upstate 06-570, 1:200), and anti-TubulinßIII (TUJ; Covance PRB-435P, 1:150). The next day, sections were washed in PBS three times 10 min each, and then incubated in secondary antibody for 2 h at room temperature. Alexa Fluor Goat α Rabbit IgG λ488 (Invitrogen A11008) and Alexa Fluor Goat α Mouse IgG λ546 (Invitrogen A 11003) were used at 1:1,000 dilution. Sections were washed again in PBS three times 10 min each, and then mounted in Vectashield mounting medium with DAPI (VECTOR H-1200). Anti-BrdU staining was conducted on embryos from pregnant females who were IP injected with BrdU (Sigma B5002, 150 μg/g) following a 2–4 h BrdU incorporation period. TUNEL staining was performed using the DeadEnd Fluorometric TUNEL System Kit (Promega G3250) as directed.
Microarray and Quantitative Real-Time PCR
Cortex, midbrain and cerebellum were dissected from brains of freshly sacrificed E17.5 embryos. Total RNA was isolated from these brain areas using RNeasy Mini Kit (Qiagen 74134) with DNaseI digestion (Invitrogen 18068-015) performed post-RNA extraction. Quality of RNA was checked and cRNA was produced by UC Davis Gene Expression Analysis facility for hybridization to Sentrix Mouse Ref-8 Expression microarray. qPCR was performed on total RNA from the same batch sent off for array. Samples were processed using Express SYBR Green qPCR Supermix (Invitrogen A10314) according to manufacture protocol. All qPCR data were generated using ß-Actin as reference (house-keeping) gene. This gene was chosen based on microarray results, which showed no significant expression variation between control and double knockout brains. Primer efficiency was tested by generating standard curves for all primers against all brain areas. Cycling was conducted as follows: pre-incubation at 95°C for 5 min, 45 cycles of amplification at 95°C for 10 s, 60°C for 20 s, and 72°C for 30 s, followed by one cycle of melting curve at 95°C for 5 s, 65°C for 1 min, and 97°C continuous.
Results
Double Knockout of c- and N-myc in NSC Severely Disrupts Brain and Cerebellar Growth, Resulting in Lethality as Early as E17.5
Previous separate single conditional targeted disruption of either c- or N-myc in NSC using nestin-cre caused varying degrees of impaired brain growth manifesting as microcephaly, however even in the case of N-myc, which had a >40% reduction in brain growth, there was no lethality apparent [16]. In those studies, an underrepresentation of N-myc flox/flox nestin-cre (NSC specific N-myc knockout) mice was present in breedings of N-myc flox/flox X N-myc flox/WT nestin-cre mice, but equally underrepresented were N-myc flox/WT cre-negative (healthy, non-phenotypic) offspring, indicating that there was likely a genetic linkage between the N-myc WT allele and the nestin-cre transgene [16], which caused these non-Mendelian, yet consistently observed ratios. Consistent with the theory of linkage, both N-myc and the nestin-cre transgene have been mapped on mouse chromosome 12, but quite some distance apart (Roussel M. and Zindy, F., unpublished observation).
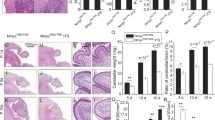
In our current DKO studies, when we did an equivalent breeding (bred N-myc flox/flox; c-myc flox/flox (doubly homozygously floxed) mice with N-myc flox/WT; c-myc flox/flox nestin-cre mice) we also observed evidence of this linkage, with a parallel underrepresentation of N-myc flox/WT; c-myc flox/flox cre-negative (healthy controls) and doubly homozygously floxed nestin-cre (DKO) mice. This was evident at some early embryonic stages examined (Fig. 1a), where only 1/6 mice (instead of one fourth) were N-myc flox/WT; c-myc flox/flox, and only 1/6 were N-myc flox/flox; c-myc flox/flox nestin-cre+. However, by age E17.5 doubly homozygously floxed nestin-cre embryos (DKO) began to be underrepresented even compared to N-myc flox/WT; c-myc flox/flox control embryos, with which they were earlier balanced in numbers. This specific underrepresentation was by ratios of approximately 2-fold or more indicating lethality, which became progressively more severe with age. By P21, there was a 2.5-fold underrepresentation of DKO compared with N-myc flox/WT; c-myc flox/flox control embryos (Fig. 1). By 4–6 weeks of age, only extraordinarily rare DKO animals survived (<5%), while N-myc flox/WT; c-myc flox/flox cre-negative mice were surviving normally and appeared healthy. After birth, surviving DKO animals were outwardly small and had severe ataxia.
Myc DKO results in lethality beginning as early as E17.5 and impaired brain growth. a Ratios of the four genotypes of embryos or pups of the indicated age produced from N-myc fl/fl; c-myc fl/fl mice crossed with N-myc fl/wt; c-myc fl/fl nestin-cre+ mice. Note a genetic linkage between the WT N-myc allele and nestin-cre transgene results in an approximately 2-fold underrepresentation of N-myc fl/wt c-myc fl/fl Cre− (control) and N-myc fl/fl c-myc fl/fl Cre+ (DKO) animals independent of phenotype. DKOs are further underrepresented from E17.5 onward indicating lethality. Numbers above bars are absolute numbers of mice produced, italicized numbers above horizontal bars are the expected number based on Mendelian ratios, and numbers within bars are% of total pups/embryos. b Brain mass as a percentage of WT. Asterisk Cre+
Brain growth was severely impaired in the DKO mice, likely explaining their lethality. The few DKO animals surviving to maturity had a 60% reduction in total brain growth and a near complete absence of cerebellar growth (Fig. 1b). Mice heterozygous for c-myc, but homozygously null for N-myc were not produced in the breeding in Fig. 1a, however they exhibited a phenotype almost as severe with a 53% reduction in brain growth. Surprisingly, mice heterozygous for N-myc, but homozygously null for c-myc had a brain growth phenotype only slightly more severe than homozygously null c-myc animals with WT N-myc (24% vs 18% reductions in brain growth), indicating that knocking out one allele of N-myc has little effect even in the context of absent c-myc (Fig. 1b).
Loss of c- and N-myc Strongly Impairs Early Cerebellar Development
At E17.5, cerebellar growth was strongly impaired in the DKO (Fig. 2a) as was overall brain growth (not shown). By P6, the cerebellar phenotype became more severe (Fig. 2b). DKO cerebellar size at P6 was only a fewfold increased compared with the DKO at E17.5 indicating minimal cerebellar growth and development occurs in the DKO during that approximately 8 day period of time. The morphology of the E17.5 and P6 DKO cerebella were also quite similar. While the DKO cerebella did grow from E17.5-P6 moderately, it is notable that the thickness of the EGL did not significantly change. In contrast, in the control, the EGL became substantially thicker. These data indicate that loss of c- and N-myc almost completely impairs EGL thickening.
The myc DKO phenotypes in the cerebellum at E17.5 and P6 (Fig. 2a) were far more severe than that observed with either single KO alone [16, 17]. These data indicate that both c- and N-myc are essential for NSC function that directs cerebellar genesis, or that N-myc is the central player but its function can be compensated for by c-myc. Interestingly, despite the apparent lethality of the nestin-cre Myc DKO and the severe cerebellar phenotype, no elevation of apoptosis was evident in the E17.5 in the cerebellum (Electronic supplementary material, Fig. S1) or brain as a whole (data shown). Therefore, c- and N-myc are not essential for NSC survival.
myc Family Expression Patterns in the Embryonic Brain Correlate with the General Domain Specificity of myc Knockout Phenotypes
We have previously observed high levels of N-Myc protein expression in the developing cortex and cerebella [16]. These findings correlated with myc family expression data at E15.5 from the brain gene expression website: http://www.stjudebgem.org/web/mainPage/mainPage.php. While c-myc expression at the protein or RNA levels is difficult to detect, there are hints of low-level expression in a number of regions including the developing cortex. N-myc is fairly ubiquitously expressed, but with relatively lower or absent expression in some midbrain regions and very high levels in the neocortex and the EGL. L-myc, despite no reported KO phenotype, has very high expression in the VZ of the embryonic midbrain, as well as moderate levels in the developing cerebellum and cortex.
A Lack of c- and N-myc Almost Completely Ablates Cerebellar Development from P6-P21
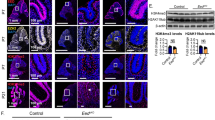
To explore the effects of the myc DKO on cerebellar development further, we examined the mature DKO cerebellum. Between P6 and P21, the DKO cerebellum grew little and there remained only one possible folium, hardly recognizable as such. Reflective of little post-natal cerebellar change in the DKO, a putative, residual EGL was evident in the P21 DKO by DAPI staining (Fig. 3 blue arrows). Staining of the DKO with NeuN, which highlights cerebellar granule neurons, indicated an absence of the internal granule cell layer (IGL) with a scattered pattern of very few of granule neurons, abnormally intense NeuN nuclear staining and possible reductions in nuclear size (Fig. 3a). The astroglia and oligodendrocytes that normally are present within the granule cell layer are ectopic and disorganized in the DKO (Fig. 3a, b), with an overrepresentation of astroglia and abnormal oligodendrocyte morphology. Purkinje neurons, which normally are present at an enormous underrepresentation compared to granule neurons, are also strikingly overrepresented in the DKO (Fig. 3a versus b, BTUJ versus NeuN staining). This is confirmed by Calbindin staining where a significant fraction of total cells in the DKO cerebellum at P21 appear to be Purkinje neurons (Fig. 4a).
Loss of c- and N-myc severely disrupts cerebellar development. Control and DKO Sagittal cerebellar sections from mature animals stained for DAPI (DNA; blue) or the indicated marker: a NeuN (predominantly granule neurons) and GFAP (astroglia), b CNPase (oligodendrocytes), and TUJ (BTUBIII; predominantly Purkinje neurons). Blue arrows in (a, b) points toward DAPI stained residual, abnormal EGL. Images were taken at 20× and scale bars = 100 μM. Note that the control images are from the tip of just one folium, whereas the DKO images are nearly the entire cerebellum
Evidence of Arrested and Impaired Development in the Mature DKO Cerebellum
While GAD67+ inhibitory neurons appear to be present in the DKO, their location, staining, and morphology are all very abnormal (Fig. 4a), suggesting a failure to mature normally. Similarly, although c- and N-myc are not strictly required for Purkinje neuron generation, nonetheless the DKO Purkinje neurons exhibited evidence of impaired maturity. The Purkinje neurons present are scattered and are quite abnormal in morphology (Fig. 3, BTUJ and Fig. 4a, Calbindin staining). While staining positive for both TUJ and Calbindin, the staining of DKO Purkinje neurons is abnormally intense. The Purkinje neurons appear to be disconnected and do not form a visible neural network. There is an absence of the normal beautiful arborization, very evident in the control (Electronic supplementary material, Fig. S2). It is unclear if these Myc-deficient Purkinje neurons possess more than rudimentary axons and dendrites, suggesting that although they are present their development is impaired. The DKO cerebellum has a severely attenuated, Math1+ EGL at E17 (Fig. 4b).
By P6, the DKO EGL is NeuN Positive and as Early as E17.5 is Mostly Math1 Negative
To explore the events earlier in cerebellar development that resulted in the severe DKO phenotype at maturity, we examined the EGL at P6 (Fig. 4b). During this stage of cerebellar development, the EGL normally consists of Math1 positive, NeuN negative CGNPs. Normally, many of the CGNPs have also begun to migrate inward to form a Math1 negative, NeuN-positive developing IGL. The findings from our staining for Math1 and NeuN in control (doubly homozygously floxed, cre negative) sections were consistent with this pattern (Fig. 5b; not shown). In striking contrast, in the DKO we observed a phenotype where the putative EGL has taken on an IGL-like fate, staining negative for Math1 and positive for NeuN (arrows in DKO panels in Fig. 5b; not shown). While the DKO has an appropriately internally localized IGL consisting of strongly NeuN-positive cells, it is highly disorganized and rostrally no longer forms an actual layer. Furthermore, the DKO cells located where the EGL should normally be are moderately positive for NeuN as well. Therefore, Myc deficiency alters CGNP fate, leading to a loss of CGNP identity, likely explaining the residual EGL apparent at P21 in the DKO (Figs. 3 and 4a). To our knowledge this is the first demonstration of an IGL-like phenotype in the EGL of a KO mouse, suggesting Myc is essential for maintaining CGNP identity and blocking differentiation. As early as E17.5, the EGL in the DKO is nearly entirely lacking Math1 positive cells, consistent with the notion of a loss progenitor cell identity (Fig. 4c).
Myc DKO decreases progenitor cell cycling at P6. a Control and DKO E17.5 cerebella were stained with the indicated markers. Green is phosphoH3; pH 3, a mitotic marker. Red is BrdU, incorporated in S phase cells. Blue is DAPI. Images in a were taken at 20× and scale bars = 100 μM. b Staining for p27KIP1(green). Yellow and white arrows indicate neuroepithelium and EGL, respectively. Bars = 200 μM. c Control and DKO E17.5 sagittal cerebellar sections were stained with nucleolin (green), a marker of cell metbolism. Images were taken at 40× and scale bars = 100 μM
c- and N-Myc are essential for CGNP cell cycling and metabolism
To examine cell cycle characteristics of control and Myc DKO cerebellar precursors, we stained E17 sections with anti-phosphoH3 antibody, a marker of mitosis, as well as anti-BrdU antibody following BrdU labeling (Fig. 5a). DKO cerebellar exhibited no clear change in mitotic labeling cells in the rhombic lip, neuroepithelium and EGL. In contrast, all germinal regions of the DKO showed a near complete absence of S phase cells as determined by BrdU staining, while control neuroepithelium, rhombic lip, and EGL exhibited a very high proportion of S phase cells following a 2 h label, reflective of the established very high proliferative rate in cerebellar precursors at this stage. The absence of a clear change in M phase cells together with a complete loss of S phase cells suggests the few DKO cells that are cycling having difficulty transiting M phase and there may be an M phase block. To analyze the cell cycle further at P6, p27KIP1 staining was conducted (Fig. 5b). The control P6 cerebellum has a handful of weakly p27-positive cells including a few in the EGL, but none in the neuroepithelium. The DKO cerebellum shows a striking induction of p27 throughout both the EGL (white arrow) and the neuroepithelium (yellow arrow). These data indicate that almost all DKO CGNPs and NSC are strongly p27 positive, consistent with the cell cycle exit suggested by the BrdU staining.
Myc has been previously linked to maintenance of cell metabolism, leading us to examine whether the DKO affects cerebellar precursor metabolism. To study potential metabolic differences, we stained with antibody against the established metabolic marker, nucleolin (Fig. 5c). The DKO cells in the EGL and in the interior of the cerebellum exhibited a pronounced decrease in nucleolin staining, reflecting impaired protein metabolism in the absence of c- and N-myc. The decreased cellular metabolism in the DKO fits with the disrupted cerebellar growth exhibited in the DKO.
Discussion
N-myc is required for normal murine and human brain growth [16, 40]. Mutation of N-myc in humans causes FS with its very large range of birth defects in a host of tissues [41]. Interestingly the range of defects in constitutive N-myc KO mice are remarkably similar to that of FS [42]. While these findings suggest a conserved role for N-myc in both human and mouse NSC that drives brain growth, the mechanisms have remained somewhat unclear. Complicating KO studies in mice has been the continued presence of c-myc, which has increased expression in the N-myc KO cerebellum and appears to compensate for the loss of N-myc [36]. Our studies here using a DKO overcome this problem, and point toward a key role for Myc in maintaining essential cerebellar progenitor cell characteristics: a specific cell cycle program, high metabolism, and inhibited differentiation (i.e., maintained pluripotency). In the Myc DKO mice, progenitors in the cerebellum lose all these properties. The DKO behaves akin to a developmental switch turning off, telling the cerebellum, via its progenitor cells that its developmental period is largely over as early as E17.5 and more strongly by P6. Interestingly, at least at the developmental stages examined, there was no clear effect of the DKO on survival.
One of the most striking features of the DKO is the changes that occur in the EGL. At P6 the EGL normal consists of CGNPs that are Math1+/NeuN−/BrdU+/p27−. An IGL is already forming that consists of cells derived from CGNPs that are almost the opposite, (e.g., Math1−/NeuN+), that have migrated inward. In the DKO at P6, the EGL has a phenotype very similar to that of the IGL, consisting of cells that are Math1−/NeuN+/BrdU−/p27+. However, like normal CGNPs of the EGL, these DKO cells still reside in the EGL domain and form a multicell layer on the outside of the developing cerebellum. They also consist of cells with small, DAPI bright nuclei, a normal feature of CGNPs in the EGL. Therefore, our data together suggest that in the absence of c- and N-myc, the DKO CGNPs have an aberrant cell fate. Instead of migrating inward, differentiating, exiting the cell cycle, and remodeling their nucleus into a larger, more euchromatic structure as normally occurs, the DKO CGNPs mostly fail to migrate and undergo an abnormal kind of differentiation right within the EGL, where they also exit the cell cycle. The fact that they nonetheless retain their location, layer structure, nuclear morphology and DAPI staining suggests that at one point they were CGNPs.
Other evidence of abnormal cell fate in the DKO comes from the Purkinje neurons that are present in the mature cerebellum. While they are overrepresented relative to granule neurons, indicating that c- and N-myc are not absolutely required for a Purkinje cell fate, the DKO Purkinje neurons are abnormal in morphology. They lack normal arborization and appear disconnected from the cerebellar neural network. This may be a cell autonomous phenotype due to loss of Myc in Purkinje progenitors, or alternatively, maybe due to the absence of granule neurons/migrating CGNPs that normal provide instructive signals for Purkinje development. In the N-myc single nestin-cre-driven KO cerebellum, Purkinje neurons are not overrepresented and despite decreased granule neurons, the Purkinje neurons still appear normal in morphology and connectedness. Therefore, the abnormal Purkinje maturation is unique to the DKO.
The effects of loss of Myc on the cell cycle of cerebellar progenitors are notable. While it is not surprising that the loss of c- and N-myc leads to the near complete absence of S phase cells, the presence of normal or near normal numbers of M phase cells is unexpected. In the previous N-myc single KOs driven by nestin-cre, decreases in both S and M phase cell were observed. The fact that in the DKO, there are almost no S phase cells but are relatively normal numbers of M phase cells suggests the DKO progenitors are blocked in M phase. This notion is supported by expression microarray experiments in which a host of M phase genes are downregulated in the DKO cerebellum [43]. Since very few DKO progenitors were in S phase, it is tempting to speculate that those few “cycling” cells are mostly stuck in M phase, and that Myc is essential for normal mitosis.
Further insight into Myc regulation of cerebellar development would be gained by exploring the potential role of the third main myc gene, L-myc, which is highly expressed in the EGL. Although constitutive L-myc KO mice were reported to have no phenotype of any kind, it seems probable that the high L-myc levels in the EGL reflect a function obscured in the L-myc KO because of the continued presence of very high levels of N-myc. Because Purkinje neurons are overrepresented in the DKO, we predict that L-myc plays a role in Purkinje neuron development as well.
Conclusion
Overall our data indicate that N- and c-myc play particularly important roles in cerebellar development. The functions of c- and N-Myc proteins in the cerebellum include maintaining pluripotency, metabolism, and cell cycling of progenitors.
References
Garson JA, Pemberton LF, Sheppard PW, Varndell IM, Coakham HB, Kemshead JT. N-myc gene expression and oncoprotein characterisation in medulloblastoma. Br J Cancer. 1989;59:889–94.
Bigner SH, Friedman HS, Vogelstein B, Oakes WJ, Bigner DD. Amplification of the c-myc gene in human medulloblastoma cell lines and xenografts. Cancer Res. 1990;50:2347–50.
MacGregar DN, Ziff EB. Elevated c-myc expression in childhood medulloblastomas. Pediatr Res. 1990;28:63–8.
Tomlinson FH, Jenkins RB, Scheithauer BW, Keelan PA, Ritland S, Parisi JE, et al. Aggressive medulloblastoma with high-level N-myc amplification. Mayo Clin Proc. 1994;69:359–65.
Bruggers CS, Tai KF, Murdock T, Sivak L, Le K, Perkins SL, et al. Expression of the c-Myc protein in childhood medulloblastoma. J Pediatr Hematol Oncol. 1998;20:18–25.
Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, et al. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26:1666–78.
Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–90.
Orian A, Grewal SS, Knoepfler PS, Edgar BA, Parkhurst SM, Eisenman RN. Genomic binding and transcriptional regulation by the Drosophila Myc and Mnt transcription factors. Cold Spring Harb Symp Quant Biol. 2005;70:1–10.
Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–70.
Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–34.
Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605.
Cawley S, Bekiranov S, Ng HH, Kapranov P, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509.
Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS ONE. 2008;3:e3650.
Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992;6:2235–47.
Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygous and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–82.
Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–712.
Hatton BA, Knoepfler PS, Kenney AM, Rowitch DH, de Alboran IM, Olson JM, et al. N-myc is an essential downstream effector of Shh signaling during both normal and neoplastic cerebellar growth. Cancer Res. 2006;66:8655–61.
Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–63.
Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–24.
Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008;2:18–21.
Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–7.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76.
Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:260–2.
Yamanaka S, Takahashi K. Induction of pluripotent stem cells from mouse fibroblast cultures. Tanpakushitsu Kakusan Koso. 2006;51:2346–51.
Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6.
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20.
Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2007;26(1):106–6.
Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–77.
Cotterman R, Knoepfler PS. N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b. PLoS ONE. 2009;4(6):e5799.
Knoepfler PS, Kenney AM. Neural precursor cycling at sonic speed: N-Myc pedals, GSK-3 brakes. Cell Cycle. 2006;5:47–52.
Hatton KS, Mahon K, Chin L, Chiu FC, Lee HW, Peng D, et al. Expression and activity of L-myc in normal mouse development. Mol Cell Biol. 1996;16:1794–804.
Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, et al. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1993;6:2248–57.
Malynn BA, de Alboran IM, O'Hagen RC, Bronson R, Davidson L, DePinho RA, et al. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–9.
Hatton B, Knoepfler P, Kenney A, Rowitch D, de Alboran I, Olson J, et al. N-myc is an essential downstream effector of Shh signaling both during normal and neoplastic cerebellar growth. Cancer Res. 2006;66:1–7.
Nagao M, Campbell K, Burns K, Kuan CY, Trumpp A, Nakafuku M. Coordinated control of self-renewal and differentiation of neural stem cells by Myc and the p19ARF-p53 pathway. J Cell Biol. 2008;183:1243–57.
Zindy F, Knoepfler PS, Xie S, Sherr CJ, Eisenman RN, Roussel MF. N-Myc and the cyclin-dependent kinase inhibitors p18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proc Natl Acad Sci USA. 2006;103:11579–83.
Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28.
Zhao X, Da D, Lim WK, Brahmachary M, Carro MS, Ludwig T, et al. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell. 2009;17:210–21.
Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–53.
van Bokhoven H, Celli J, van Reeuwijk J, Rinne T, Glaudemans B, van Beusekom E, et al. MYCN haploinsufficiency is associated with reduced brain size and intestinal atresias in Feingold syndrome. Nat Genet. 2005;37:465–7.
Teszas A, Meijer R, Scheffer H, Gyuris P, Kosztolanyi G, van Bokhoven H, et al. Expanding the clinical spectrum of MYCN-related Feingold syndrome. Am J Med Genet A. 2006;140:2254–6.
Marcelis CL, Hol FA, Graham GE, Rieu PN, Kellermayer R, Meijer RP, et al. Genotype-phenotype correlations in MYCN-related Feingold syndrome. Hum Mutat. 2008;29:1125–32.
Wey A, Knoepfler PS. c-myc and N-myc promote active stem cell metabolism and cycling as architects of the developing brain. Onco Target. 2010;1:1–10.
Acknowledgements
This work was supported by the following grants to PK: NIH Howard Temin Award K01 CA114400, A Shriners Hospital for Children grant, and the Basil O’Connor Starter Scholar Award from the March of Dimes.
Conflicts of Interest
The authors state they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 6604 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Wey, A., Cerdeno, V.M., Pleasure, D. et al. c- and N-myc Regulate Neural Precursor Cell Fate, Cell Cycle, and Metabolism to Direct Cerebellar Development. Cerebellum 9, 537–547 (2010). https://doi.org/10.1007/s12311-010-0190-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12311-010-0190-9