Abstract
Multiple myeloma (MM) is an incurable disease, however, novel therapeutic agents has significantly improved its prognosis. In this study we analyzed if polymorphisms in the genes of β-catenin and glutathione-S-transferase have affected the clinical course, treatment response and progression-free survival (PFS) of MM patients. Ninety-seven MM patients were involved who were administered immunomodulatory drug (Imid) or alkylating agent-based therapy. β-catenin (CTNNB1, rs4135385 A > G, rs4533622 A > C) and glutathione-S-transferase (GSTP1 105, GSTP1 114) gene polymorphisms were analyzed by Light SNiP assays. The distribution of CTNNB1 (rs4135385) AA, AG and GG genotypes were 48.4%, 47.4% and 4,1%, respectively. Patients with AA genotype were older than those who carried G allele (64.5 vs. 61.0 years of age, p < 0.05). Response to Imid-based therapies (p < 0.05) and PFS (p = 0.032) were significantly more favourable in the AA homozygous group. The other polymorphism (rs4533622) of β-catenin gene did not markedly influence these clinical parameters, although MM was diagnosed at significantly younger age in subjects with CC genotype compared to AG/AA combined genotypes (59.1 vs. 65.7 years, p = 0.015). When GSTP1 polymorphisms were investigated, no such significant associations were observed. Our results demonstrate that the polymorphism of β-catenin gene (rs4135385) may be an independent predictive factor in MM.
Similar content being viewed by others
Introduction
Multiple myeloma (MM) is a malignant hematologic disorder that accounts for 1 % of all cancer cases. It is considered as an incurable disease however there has been a significant improvement in patients’ survival, thanks to the novel drugs that have been recently introduced. Subclonal heterogeneity of MM has been described by several studies. The heterogenous molecular pattern of MM results in different therapeutic responses [1]. Due to molecular heterogeneity, there is a need to identify new biomarkers of drug resistance. Biomarker tests can improve the efficacy of traditional therapies, therefore determination of molecular profile in cancer patients is becoming standard of care [2]. In multiple myeloma, new therapies include immunomodulatory agents (IMIDs), proteasome inhibitors (PI) and monoclonal antibodies [3]. Thalidomide, lenalidomide and pomalidomide are immunomodulatory drugs that can be administered in first-line treatment, maintenance and/or relapse settings of MM. Imids act via the cereblon-β-catenin pathway. Cereblon (CRBN) is expressed in several malignant cell lines, such as myeloma and lymphoma cells. It is a part of the CRL4 complex that has a major role in the ubiquitination of IKZF1 and IKZF3 transcription factors [4, 5]. Thalidomide binds to cereblon and inhibits its E3 ubiquitin ligase function. Moreover, thalidomide facilitates upregulation of p21 and downregulation of interleukine-8, that results in an arrest in the G0/G1 transition of the cell cycle [6, 7]. However, myeloma cells are able to become resistant against Imids which process is still not clear in details. Lately, the pathological regulation of β-catenin/Wnt pathway has been described in the lenalidomide resistance setting. β-catenin plays a key role in the Wnt nuclear transcription pathway. It is phosphorylated by the Axin/CK1a/APC/GSK3 complex, then it is transported to the nucleus, which results in an improvement of the survival rate of tumor cells. Lenalidomide and thalidomide increase the intracellular concentration of β-catenin, but also facilitate the expression of c-myc and other anti-apoptotic factors, and treatment may become ineffective consequently. Overall, cereblon is responsible for the acitivity of Imids – thalidomide and lenalidomide -, but the Wnt/β-catenin pathway may contribute to the development of resistance against these drugs [8,9,10]. Genes that encode cereblon (CRBN) and β-catenin (CTNBB1) can be found on chromosome 3 in humans. The exchange of adenine and guanine in intron 13 is responsible for rs4135385 polymorphism of CTNBB1 gene, while rs4533622 polymorphism incorporates an adenine-citozine substitution [4, 5]. CTNBB1 (rs4533622) polymorphism was reported to play a role in the clinical course of MM, on the other hand, rs4135385 polymorphism can be a predisposing factor at the onset of the disease. Cyclophosphamide – thalidomide – dexamethasone (CTD) therapy was found to be more effective and less progression was observed in CTNBB1 (rs4135385) AA homozygous patients. Patients carrying the CTNNB1 (rs4533622) AA genotype were better responders to the first-line therapy with thalidomide-containing regimens. These results may highlight the need for administering personalized treatment for MM patients [11, 12].
The polymorphisms of some other drug metabolizing enzyme genes may also influence treatment efficacy in MM patients. For instance, glutathione-S-transferase (GST) enzymes are responsible for the detoxification of several xenobiotics and chemotherapeutical agents [13]. GSTP1 enzyme is an important member of the GST family, as it metabolizes melphalan, cyclophosphamide, vincristine, doxorubicine, cisplatina, etoposide and chlorambucil. Environmental carcinogens, such as benzene, dioxins, and fume are also the substrates of GSTP1 [14]. Aminoacid substitutions of Ile105Val and Ala114Val are likely to have a significance in the modulation of enzyme activity [15]. In patients having GSTP1 105 variant genotype, detoxification process was less active, therefore chemotherapeutical agents can be more effective. In MM patients carrying GSTP1 105Ile homozygous genes, progression-free (PFS) and overall survival results were reported to be more favourable [16].
Our aim was to investigate if CRBN, CTNNB1 and GSPT1 gene polymorphisms have any effect on treatment response and survival data of a large group of Hungarian MM patients and if administration of personalized treatment can be considered in this disease.
Patients and Methods
Clinical Data
The clinical files of multiple myeloma patients were reviewed with particular reference to age, sex, clinical stage, response to treatment and survival. ISS stages were determined using the International Myeloma Working Group criteria. FISH results of unfavourable prognosis included t(4;14), t(14;16) and del(17p). Examining the survival rates, overall survival (OS) was determined by consideration of death events due to any reasons, while progression-free survival (PFS) was determined by consideration of relapses, deaths or disease progression that indicated further treatment. Descriptive statistical analysis was used to characterize the patient populations. Normality of the parameters were examined applying the Wilk-Saphiro test. Comparing two groups, F probe and t test were administered by normal distribution of the parameters, otherwise the non-parametrical Mann-Whitney test was applied. Differences were significant if probability level was less than 5% (p < 0.05). Survival rates were calculated using the Kaplan-Meier’s method, while the survival data were compared using the log-rank test. Cox uni- and multivariable regression analysis were used to assess several risk factors simultaneously in terms of survival data.
Determination of Gene Polymorphisms
Genomic DNA Extraction
DNA for genotyping was extracted from peripheral blood samples obtained into K3-EDTA Vacutainer tubes (Becton Dickinson, San Jose, CA, USA) using QiaAmp DNA Blood Mini Kit (Qiagen GmbH, Germany) according to the manufacturer’s recommendations.
Genotyping of the CRBN, CTNNB1 and GSTP1 Gene Polymorphisms
LightSNip typing assays were applied to determine CRBN (rs121918368C > T), CTNNB1 polymorphisms (rs4135385 A > G, rs4533622 A > C) and GSTP1105 (rs1695A > G) and 114 (rs1138272C > T) (TIB-MolBiol, Berlin, Germany) on a LightCycler 480 Real-time PCR Instrument (Roche Diagnostics, Mannheim, Germany). Amplifications were performed based on the recommendations of the manufacturer. PCR reaction was carried out in 20 μl volume containing the LightCyclerFastStart DNA Master HybProbe (Roche Diagnostics). Samples were run in duplicates.
Results
Ninety-seven multiple myeloma patients were involved in the study who were treated in our institution between January 2012 and December 2016. Their mean age was 62.47 years at diagnosis, male-female ratio was 43:54. The distribution of ISS1, 2 and 3 stages were 27.8%, 42.3% and 29.9%, respectively. FISH tests were performed in 67 patients and 42 (62.7%) of them had a result of unfavourable prognosis. Treatment modalities included thalidomide-based (59 cases), lenalidomide-based (18 cases), and alkylating-agent based (57 cases) regimens. Thirty-eight patients received two or more treatment lines.
In this cohort, the distribution of CTNNB1 (rs4135385) AA, AG and GG genotypes were 48.5%, 47.4% and 4.1%, respectively. Patients with AA genotype were older than those who carried G allele (64.5 vs. 61.0 years of age, p = 0.05). However, the presence of the CTNNB1 (rs4135385) AA genotype did not show association with ISS stage or FISH test results (Table 1). Regarding the rs4533622 polymorphism of the CTNNB1, the different genotypes were detected in the following ratio: AA genotype in 18.5%, AC in 50.5% and CC in 31% of these patients. MM was diagnosed at significantly younger age in those carrying the CC genotype (59.1 vs. 65.7 years, p = 0.015). Additionally, females were more likely to bear the AA genotype (Table 2). Similarly to the other polymorphism of β-catenin, there was no relationship between allele distribution and clinical stages or FISH results. In terms of the CRBN (rs121918368) polymorphism, all patients carried the CC genotype, thus, no further analysis was performed in regard to clinical features, treatment results and survival ratio.
For the GSTP1 105 polymorphism, there were 54 (55.7%) patients carried AA (Ile/Ile), 33 (34%) had AG (Ile/Val) and 10 (10.3%) had GG (Val/Val) genotype. When GSTP1 114 polymorphism was tested among these MM individuals, CC (Ala/Ala) genotype was found in 85 (87.6%) subjects, while CT (Ala/Val) genotype was found in 12 (12.4%) patients. No association was observed between these polymorphic variants in GSTP1 and demographical data.
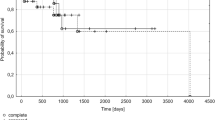
We also investigated the effect of CTNNB1, CRBN and GSTP1 polymorphisms on response to chemotherapy. We found that response to Imids-based therapies (p < 0.05) and PFS (p = 0.032) were significantly more favourable in AA homozygous group (Table 1, Fig. 1). Cox univariable (HR: 2.371 [1.026–5.477], P = 0.043), and multivariable analysis tests (HR: 2.554 [1.099–5.933], P = 0.029) for PFS proved that CTNNB1 (rs4135385) polymorphism can be considered as an independent prognostic factor besides ISS stages and FISH results (Tables 3 and 4). In patients with stage II and stage III disease, G carriers were expected to have more unfavourable survival results than AA homozygous individuals (Table 5). In contrast, the other polymorphism (rs4533622) of β-catenin gene did not markedly influence the effectivity of thalidomide and lenalidomide-based therapies (Table 1). Progression-free survival was markedly favourable in the AA group; however, the survival curves did not differ significantly from each other (Fig. 2). β-catenin gene polymorphisms did not have any impact on the occurrence of common side effects of the Imid-based therapies (Tables 1 and 2).
Interestingly, when GSTP1 105 and 114 polymorphisms were investigated, no significant association was observed between the response to chemotherapy or survival ratio of these MM patients.
Discussion
Recently, it has been becoming a trend to administer personalized treatment modalities in cancer patients to improve survival data. Molecular profiling can be a good tool for effective personalized care [2]. In our study, the gene polymorphisms of the cereblon-beta-catenin pathway and glutathione-S-tranferase were investigated in multiple myeloma patients. Several publications have highlighted the importance of CRBN gene expression in the effectivity of Imid-based therapies. Thalidomide resistance may occur in case of a low CRBN activity, while high levels of CRBN mRNA were found to be associated with better responses to thalidomide therapy [17, 18]. Our patient population was homogenous in terms of the CRBN gene polymorphism as each of them carried the CC genotype. On the other hand, we could find some links with the CTNNB1 (rs4533622) polymorphism. In CC homozygous patients, the onset of the disease occurred at a significantly younger age and A alleles were significantly less common in males. These associations have never been published as yet. CTNNB1 (rs4533622) polymorphism was reported to influence the ISS stages as patients carrying an A allele had more advanced disease. Butrym et al. also described better treatment responses and more neutropenic events in AA homozygous patients [11]. However, we could not confirm such associations in our cohort.
A few studies investigated the role of CTNNB1 (rs4135385) polymorphism in the pathogenesis of malignant disorders. In a Chinese population, the susceptibility of gastric cancer was higher in patients carrying an A allele [19]. Breast cancer was found to be more common in the AG heterozygous group [20]. On the other hand, no predisposing role of any CTNNB1 (rs4135385) polymorphism could be confirmed in terms of multiple myeloma. However, cyclophosphamide – thalidomide – dexamethasone (CTD) combination therapy was more effective in MM patients carrying the AA genotype [11]. Our results were in accordance with this finding as therapeutic responses and progression-free survival results were more favourable in AA homozygous patients after Imid-based therapies. Moreover, CTNNB1 (rs4135385) polymorphism were found to be an independent prognostic factor in terms of PFS results besides ISS stages and FISH results. No associations were detected between beta-catenin gene polymorphisms and the severity of any Imid-related side effects. Being aware of this polymorphism, personalized treatment strategies can be suggested for multiple myeloma patients.
Glutathione-S-transferase enzyme plays an important role in the metabolism of carcinogens and cytostatic agents as well. GSTP1 105 variant of this enzyme has less ability for detoxification, therefore the effectivity of cytostatic agents is increased in such multiple myeloma patients. In patients carrying the GSTP1 105Ile homozygous genotype, event-free and overall survival results were reported to be more favourable after alkylating agent or anthracyclin-based treatment [16]. In our cohort, neither GSTP1 105, nor GSTP1 114 polymorphisms had any impact on the treatment results and survival data.
Our results highlighted that beta-catenin gene rs4135385 gene polymorphism may influence the clinical features of multiple myeloma patients and have some impact on the treatment results and survival data after Imid-based therapies. Determination of molecular patterns and gene polymorphisms in multiple myeloma patients is an important aspect of personalized medicine which may improve patients’ survival. [2].
References
Jaak Ph J, Klaus S, Andreas V et al (2018) Preventive, predictive, and personalized medicine for effective and affordable cancer care. EPMA Journal 9:113–123
Raza S, Safyan RA, Rosenbaum E, Bowman AS, Lentzsch S (2017) Optimizing current and emerging therapies in multiple myeloma: a guide for the haematologist. Ther Adv Hematol 8:55–70
van Hengel J, Nollet F, Berx G, van Roy N, Speleman F, van Roy F (1995) Assignment of the human beta-catenin gene (CTNNB1) to 3p22-p21.3 by fluorescence in situ hybridization. Cytogenet Cell Genet 70:68–70
Nollet F, Berx G, Molemans F, van Roy F (1996) Genomic organization of the human beta-catenin gene (CTNNB1). Genomics. 32:413–424
Lin Y, Huang X, He X et al (2015) A novel effect of thalidomide and its analogs: supression of cereblon ubiquitination enhances ubiquitin ligase function. FASEB J 9:4829–4839
Chang XB, Stewart AK (2011) What is the functional role of thalidomid binding protein cereblon? Int J Biochem Mol Biol 2:287–294
Qiang YW, Rudikoff S (2009) Wnt signaling pathways in multiple myeloma. In: Georgiev M, Bachev E (eds) Multiple myeloma: symptoms, diagnosis and treatment. Nova Science Publisher, New York, pp 51–75
Tian J, He H, Lei G (2014) Wnt/ß-catenin pathway in bone cancers. Tumour Biol 35:9439–9445
Bjorklund C, Ma W, Wang ZQ et al (2011) Evidence of a role for activation of Wnt/ß-catenin signaling in the resistance of plasma cells to lenalidomide. J Biol Chem 286:11009–11020
Butrym A, Rybka J, Lacina P et al (2015) Polymorphisms within beta-catenin encoding gene affect multiple myeloma development and treatment. Leuk Res 39:1462–1466
Huang SY, Lin CW, Lin HH, Yao M, Tang JL, Wu SJ, Chen YC, Lu HY, Hou HA, Chen CY, Chou WC, Tsay W, Chou SJ, Tien HF (2014) Expression of cereblon protein assessed by immunohistochemical staining in myeloma cells is associated with superior response of thalidomide- and lenalidomide-based treatment, but not bortezomib-based treatment, in patients with multiple myeloma. Ann Hematol 93:1371–1380
Srivastava SK, Watkins SC, Schuetz E, Singh SV (2002) Role of glutathione conjugate efflux in cellular protection against benzo (a) pyrene-7,8-diol-9,10-epoxide-induced DNA damage. Mol Carcinog 33:156–162
Taningher M, Malacarne D, Izzotti A, Ugolini D, Parodi S (1999) Drug metabolism polymorphisms as modulators of cancer susceptibility. Mutat Res 436:227–261
Johansson AS, Stenberg G, Widersten M, Mannervik B (1998) Structure-activity relationships and thermal stability of human glutathione transferase P1-1 governed by the H-site residue 105. J Mol Biol 278:687–698
Dasgupta RK, Adamson PJ, Davies FE et al (2003) Polymorphic variation in GSTP1 modulates outcome following therapy for multiple myeloma. Blood. 102:2345–2350
Chang X, Xu Q, Hou Y et al (2017) Mouse monoclonal antibodies generated from full length human cereblon: detection of cereblon protein in patients with multiple myeloma. Int J Mol Sci 18: 1999, https://doi.org/10.3390/ijms18091999.
Gandhi AK, Mendy D, Waldman M, Chen G, Rychak E, Miller K, Gaidarova S, Ren Y, Wang M, Breider M, Carmel G, Mahmoudi A, Jackson P, Abbasian M, Cathers BE, Schafer PH, Daniel TO, Lopez-Girona A, Thakurta A, Chopra R (2014) Measuring cereblon as a biomarker of response or resistance to lenalidomide and pomalidomide requires use of standardized reagents and understanding of gene complexity. Br J Haematol 164:233–244
Wang S, Tian Y, Wu D, Zhu H, Luo D, Gong W, Zhou Y, Zhou J, Zhang Z (2012) Genetic variation of CTNNB1 gene is associated with susceptibility and prognosis of gastric cancer in a Chinese population. Mutagenesis. 27:623–630
Alanazi MS, Parine NR, Shaik JP, Alabdulkarim HA, Ajaj SA, Khan Z (2013) Association of single nucleotide polymorphisms in Wnt signaling pathway genes with breast cancer in saudi patients. PLoS One 8:e59555
Funding
Open access funding provided by University of Debrecen (DE). This work was supported by the Hungarian Hematology and Transfusiology Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no conflicts of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Pál, I., Szilágyi, B., Nagy, B. et al. The Impact of Beta-Catenin and glutathione-S-transferase Gene Polymorphisms on the Treatment Results and Survival of Multiple Myeloma Patients. Pathol. Oncol. Res. 26, 1633–1638 (2020). https://doi.org/10.1007/s12253-019-00747-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-019-00747-5