Abstract
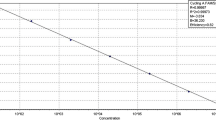
To evaluate the effective implantation of a specific protective culture of Penicillium nalgiovense, a real-time quantitative PCR (qPCR) using SYBR Green methodology was developed. Two specific primers were designed on the basis of the published partial sequences of the Internal Transcribe Spacer (ITS)1–5.8S-ITS2 region of various strains of P. nalgiovense. Using the developed method, a PCR product of 51 bp with a T m value 81.34 °C was detected. T m values of the amplified product allowed specific differentiation between P. nalgiovense and the remaining mould species tested. The developed qPCR method was tested on inoculated slices of dry-cured sausage (‘salchichón’) showing an efficiency of 97.24 %, a R 2 value of 0.99 and a detection limit of P. nalgiovense of 1 log colony-forming units (cfu)/cm2. The qPCR method demonstrated that the protective strain of P. nalgiovense grew and competed against an ochratoxin A (OTA)-producing Penicillium verrucosum strain on commercial dry-cured sausage. This qPCR method provides a specific, accurate and sensitive detection and quantification of P. nalgiovense on dry-cured sausage salchichón in order to estimate its colonization during their processing. This assay will improve strategies to prevent and control unwanted mould colonization and OTA risk in dry-cured meat commodities.



Similar content being viewed by others
References
Asefa DT, Kuré CF, Gjerde RO, Omer MK, Langsrud S, Nesbakken T, Skaar I (2010) Fungal growth pattern, sources and factors of mould contamination in a dry-cured meat production facility. Int J Food Microbiol 140:131–135. doi:10.1016/j.ijfoodmicro.2010.04.008
Battilani P, Pietri VA, Giorni P, Formenti S, Bertuzzi T, Virgili R, Kozakiewicz Z (2007) Penicillium populations in dry-cured ham manufacturing plants. J Food Prot 70:975–980
Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H (2010) ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10:189. doi:10.1186/1471-2180-10-189
Bernáldez V, Córdoba JJ, Rodríguez M, Cordero M, Polo L, Rodríguez A (2013) Effect of Penicillium nalgiovense as protective culture in processing of dry-fermented sausage “salchichón”. Food Control 32:69–76. doi:10.1016/j.foodcont.2012.11.018
Bernáldez V, Rodríguez A, Martín A, Lozano D, Córdoba JJ (2014) Development of a multiplex qPCR method for simultaneous quantification in dry-cured ham of an antifungal-peptide Penicillium chrysogenum strain used as protective culture and aflatoxin-producing moulds. Food Control 36:257–266. doi:10.1016/j.foodcont.2013.08.020
Bogs C, Battilani P, Geisen R (2006) Development of a molecular detection and differentiation system for ochratoxin A producing Penicillium species and its application to analyse the occurrence of Penicillium nordicum in cured meats. Int J Food Microbiol 107:39–47. doi:10.1016/j.ijfoodmicro.2005.08.010
Castellari C, Quadrelli AM, Laich F (2010) Surface mycobiota on Argentinean dry fermented sausages. Int J Food Microbiol 142:149–155. doi:10.1016/j.ijfoodmicro.2010.06.016
Edwards SG, O’Callaghan J, Dobson ADW (2002) PCR-based detection and quantification of mycotoxigenic fungi. Mycol Res 106:1005–1025. doi:10.1017/S0953756202006354
Fredlund E, Gidlund A, Olsen M, Börjesson T, Spliid NHH, Simonsson M (2008) Method evaluation of Fusarium DNA extraction from mycelia and wheat for downstream real-time PCR quantification and correlation to mycotoxin levels. J Microbiol Methods 73:33–40. doi:10.1016/j.mimet.2008.01.007
Gil-Serna J, González-Salgado A, González-Jaén MT, Vázquez C, Patiño B (2009) ITS-based detection and quantification of Aspergillus ochraceus and Aspergillus westerdijkiae in grapes and green coffee beans by real-time quantitative PCR. Int J Food Microbiol 131:162–167. doi:10.1016/j.ijfoodmicro.2009.02.008
González-Salgado A, Patiño B, Gil-Serna J, Vázquez C, González-Jaén MT (2009) Specific detection of Aspergillus carbonarius by SYBR® Green and TaqMan® quantitative PCR assays based on the multicopy ITS2 region of the rRNA gene. FEMS Microbiol Lett 295:57–66. doi:10.1111/j.1574-6968.2009.01578.x
Iacumin L, Chiesa L, Boscolo D, Manzano M, Cantoni C, Orlic S, Comi G (2009) Moulds and ochratoxin A on surfaces of artisanal and industrial dry sausages. Food Microbiol 26:65–70. doi:10.1016/j.fm.2008.07.006
Kuré CF, Skaar I, Brendehaug J (2004) Mould contamination in production of semi-hard cheese. Int J Food Microbiol 93:41–49. doi:10.1016/j.ijfoodmicro.2003.10.005
Le Dréan G, Mounier J, Vasseur V, Arzur D, Habrylo O, Barbier G (2010) Quantification of Penicillium camemberti and P. roqueforti mycelium by real-time PCR to assess their growth dynamics during ripening cheese. Int J Food Microbiol 138:10–17. doi:10.1016/j.ijfoodmicro.2009.12.013
López-Díaz TM, Santos JA, García-López ML, Otero A (2001) Surface mycoflora of a Spanish fermented meat sausage and toxigenicity of Penicillium isolates. Int J Food Microbiol 68:69–74. doi:10.1016/S0168-1605(01)00472-X
Lund F, Frisvad JC (2003) Penicillium verrucosum in wheat and barley indicates presence of ochratoxin A. J Appl Microbiol 95:1117–1123. doi:10.1046/j.1365-2672.2003.02076.x
Mayer Z, Bagnara A, Färber P, Geisen R (2003) Quantification of the copy number of nor-1, a gene of the aflatoxin biosynthetic pathway by real-time PCR, and its correlation to the cfu of Aspergillus flavus in foods. Int J Food Microbiol 82:143–151. doi:10.1016/S0168-1605(02)00250-7
Mulè G, Susca A, Logrieco A, Stea G, Visconti A (2006) Development of a quantitative real-time PCR assay for the detection of Aspergillus carbonarius in grapes. Int J Food Microbiol 111:S28–S34. doi:10.1016/j.ijfoodmicro.2006.03.010
Núñez F, Rodríguez MM, Bermúdez E, Córdoba JJ, Asensio MA (1996) Composition and toxigenic potential of the mould population on dry-cured Iberian ham. Int J Food Microbiol 32:185–197. doi:10.1016/0168-1605(96)01126-9
Papagianni M, Papamichael EM (2007) Modeling growth, substrate consumption and product formation of Penicillium nalgiovense grown on meat simulation medium in submerged batch culture. J Ind Microbiol Biotechnol 34:225–231. doi:10.1007/s10295-006-0190-4
Petzinger E, Ziegler K (2000) Ochratoxin A from a toxicological perspective. J Vet Pharmacol Ther 23:91–98. doi:10.1046/j.1365-2885.2000.00244.x
Postollec F, Falentin H, Pavan S, Combrisson J, Sohier D (2011) Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol 28:848–861. doi:10.1016/j.fm.2011.02.008
Ririe KM, Rasmussen RP, Wittwer CT (1997) Product differentiation by analysis of DNA melting curves during the polymerase chain reactions. Anal Biochem 245:154–160. doi:10.1006/abio.1996.9916
Rodríguez A, Luque MI, Andrade MJ, Rodríguez M, Asensio MA, Córdoba JJ (2011a) Development of real-time PCR methods to quantify patulin-producing molds in food products. Food Microbiol 28:1190–1199. doi:10.1016/j.fm.2011.04.004
Rodríguez A, Rodríguez M, Luque MI, Justesen AF, Córdoba JJ (2011b) Quantification of ochratoxin A-producing molds in food products by SYBR Green and TaqMan real-time PCR methods. Int J Food Microbiol 149:226–235. doi:10.1016/j.ijfoodmicro.2011.06.019
Rodríguez A, Córdoba JJ, Gordillo R, Córdoba MG, Rodríguez M (2012a) Development of two quantitative real-time PCR methods based on SYBR Green and TaqMan to quantify sterigmatocystin-producing molds in foods. Food Anal Methods 5:1514–1525. doi:10.1007/s12161-012-9411-9
Rodríguez A, Rodríguez M, Andrade MJ, Córdoba JJ (2012b) Development of a multiplex real-time PCR to quantify aflatoxin, ochratoxin A and patulin producing molds in foods. Int J Food Microbiol 155:10–18. doi:10.1016/j.ijfoodmicro.2012.01.007
Rodríguez A, Rodríguez M, Luque MI, Justesen AF, Córdoba JJ (2012c) A comparative study of DNA extraction methods to be used in real-time PCR based quantification of ochratoxin A-producing molds in food products. Food Control 25:666–672. doi:10.1016/j.foodcont.2011.12.010
Rodríguez A, Rodríguez M, Luque MI, Martín M, Córdoba JJ (2012d) Real-time PCR assays for detection and quantification of aflatoxin producing molds in foods. Food Microbiol 31:89–99. doi:10.1016/j.fm.2012.02.009
Rodríguez A, Rodríguez M, Martín A, Delgado J, Córdoba JJ (2012e) Presence of ochratoxin A on the surface of dry-cured Iberian ham after initial fungal growth in drying stage. Meat Sci 92:728–734. doi:10.1016/j.meatsci.2012.06.029
Rodríguez A, Rodríguez M, Martín A, Núñez F, Córdoba JJ (2012f) Evaluation of hazard of aflatoxin B1, ochratoxin A and patulin production in dry-cured ham and early detection of producing moulds by qPCR. Food Control 27:118–126. doi:10.1016/j.foodcont.2012.03.009
Sánchez B, Rodríguez M, Casado EM, Martín A, Córdoba JJ (2008) Development of an efficient fungal DNA extraction method to be used in random amplified polymorphic DNA–PCR analysis to differentiate cyclopiazonic acid mold producers. J Food Prot 71:2497–2503
Sardiñas N, Vázquez C, Gil-Serna J, González-Jaén M, Patiño B (2010) Specific detection of Aspergillus parasiticus in wheat flour using a highly sensitive PCR assay. Food Addit Contam Part A 27:853–858. doi:10.1080/19440041003645779
Sardiñas N, Vázquez C, Gil-Serna J, González-Jaén MT, Patiño B (2011) Specific detection and quantification of Aspergillus flavus and Aspergillus parasiticus in wheat flour by SYBR® Green quantitative PCR. Int J Food Microbiol 145:121–125. doi:10.1016/j.ijfoodmicro.2010.11.041
Sobhy H, Colson P (2012) Gemi: PCR primers prediction from multiple alignments. Comp Funct Genomics Article ID 783138. doi: 10.1155/2012/783138
Suanthie Y, Cousin MA, Woloshuk CP (2009) Multiplex real-time PCR for detection and quantification of mycotoxigenic Aspergillus, Penicillium and Fusarium. J Stored Prod Res 45:139–145. doi:10.1016/j.jspr.2008.12.001
Sweeney MJ, Dobson ADW (1998) Mycotoxin production by Aspergillus, Fusarium and Penicillium species. Int J Food Microbiol 43:41–158. doi:10.1016/S0168-1605(98)00112-3
White TJ, Burns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelgard DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, p 315–322
Acknowledgments
This work has been funded by the projects AGL2007-64639 and AGL2010-21623 of the Spanish Comisión Interministerial de Ciencia y Tecnología, Carnisenusa CSD2007-00016, Consolider Ingenio 2010 and GRU10162 of the Gobierno de Extremadura and FEDER. Dr. Alicia Rodríguez was the recipient of a grant from the University of Extremadura (Plan de Iniciación a la Investigación, Desarrollo Tecnológico e Innovación de la Universidad de Extremadura 2012, Becas Puente).
Conflict of Interest
M. Cordero, J.J. Córdoba, V. Bernáldez, M. Rodríguez and A. Rodríguez declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article does not contain any studies with human or animal subjects.
Rights and permissions
About this article
Cite this article
Cordero, M., Córdoba, J.J., Bernáldez, V. et al. Quantification of Penicillium nalgiovense on Dry-Cured Sausage ‘Salchichón’ Using a SYBR Green-Based Real-Time PCR. Food Anal. Methods 8, 1582–1590 (2015). https://doi.org/10.1007/s12161-014-0078-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-014-0078-2