Abstract
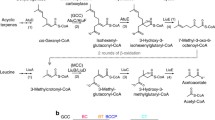
Methylcrotonyl-CoA carboxylase (MCCC) is a biotin dependent enzyme, that plays a crucial role in leucine metabolism. The enzyme comprises a biotin carboxylase (BC), a carboxyltransferase (CT), and a biotin carboxyl carrier protein (BCCP) domain. MCCC is synthesized as an apo-protein, and is posttranslationally modified at a lysine residue, conserved in the biotin carboxyl carrier protein (BCCP) domain. In order to understand the structure, function and interactions of L. major MCCC, we have expressed and characterized its domains. Here we report the complete chemical shift assignments of MCCC BCCP domain of L. major. Furthermore, we have used the assignments to generate a model of the same, using CS-Rosetta. We have also followed its chemical shift perturbations upon biotin modification. Changes were observed at the lysine 51 amide, that undergoes biotin modification, and a few others present in its immediate neighborhood.



Similar content being viewed by others
References
Chu CH, Cheng D (2007) Expression, purification, characterization of human 3-methylcorotonyl-CoA carboxylase (MCCC). Protein Expr Purif 53:421–427
Croft SL, Coombs GH (2003) Leishmaniasis-current chemotherapy and recent advances in the search for novel drugs. Trends Parasitol 19:502–508
Cronan JE (2014) Biotin and lipoic acid: synthesis attachment and regulation. EcoSal Plus. https://doi.org/10.1128/ecosalplus.3.6.3.5
Delaglio F, Grzesiek S et al (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6(3):277–293
Huang CS, Ge P et al (2011) An unanticipated architecture of the 750-kDa α6β6 holoenzyme of 3-methylcrotonyl-CoA carboxylase. Nature 481:219–223
Keller RLJ (2004) Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment dissertation. Swiss Federal Institute of Technology, Zurich
Lange OF, Rossi P et al (2012) Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci 109:10873–10878
Ponte-Sucre A, Gamarro F et al (2017) Drug resistance and treatment failure in leishmaniasis: a 21st century challenge. PLoS Negl Trop Dis 11:e0006052
Rakotomanga M, Saint-Pierre-Chazalet M et al (2005) Alteration of fatty acid and sterol metabolism in miltefosine-resistant Leishmania donovani promastigotes and consequences for drug-membrane interactions. Antimicrob Agents Chemother 49:2677–2686
Shen Y, Lange O et al (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA 105:4685–4690
Tasbihi M, Shekari F et al (2020) Comparative mitochondrial proteomics of Leishmania tropica clinical isolatesresistant and sensitive to meglumine antimoniate. Parasitol Res 119:1857–1871
Tong L (2013) Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci 70:863–891
Williamson MP (2013) Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc 73:1–16
Acknowledgements
Authors thank the Department of Biotechnology (DBT), Govt. of India for financial and infrastructure support. The following reagent was obtained through BEI Resources, NIAID, NIH as part of the Human Microbiome Project: Genomic DNA from Leishmania major, strain NIH SD (MHOM/SN/74/SD).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rajak, M.K., Sundd, M. Chemical shift assignments of the biotin carboxyl carrier protein domain of L. major Methylcrotonyl-CoA carboxylase. Biomol NMR Assign 15, 249–253 (2021). https://doi.org/10.1007/s12104-021-10013-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-021-10013-y